Francesco Iorio
Configuration Design of Mechanical Assemblies using an Estimation of Distribution Algorithm and Constraint Programming
Mar 14, 2025Abstract:A configuration design problem in mechanical engineering involves finding an optimal assembly of components and joints that realizes some desired performance criteria. Such a problem is a discrete, constrained, and black-box optimization problem. A novel method is developed to solve the problem by applying Bivariate Marginal Distribution Algorithm (BMDA) and constraint programming (CP). BMDA is a type of Estimation of Distribution Algorithm (EDA) that exploits the dependency knowledge learned between design variables without requiring too many fitness evaluations, which tend to be expensive for the current application. BMDA is extended with adaptive chi-square testing to identify dependencies and Gibbs sampling to generate new solutions. Also, repair operations based on CP are used to deal with infeasible solutions found during search. The method is applied to a vehicle suspension design problem and is found to be more effective in converging to good solutions than a genetic algorithm and other EDAs. These contributions are significant steps towards solving the difficult problem of configuration design in mechanical engineering with evolutionary computation.
Machine learning prediction of cancer cell sensitivity to drugs based on genomic and chemical properties
Mar 18, 2013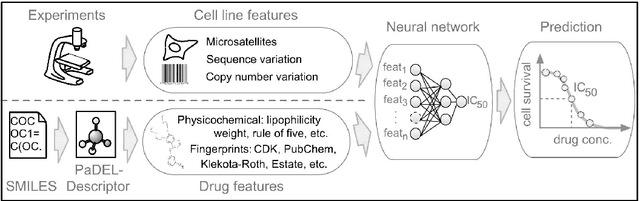
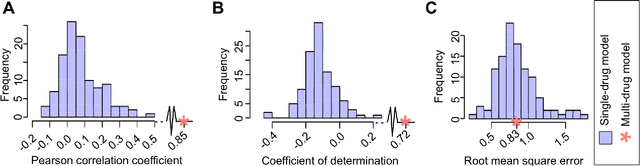
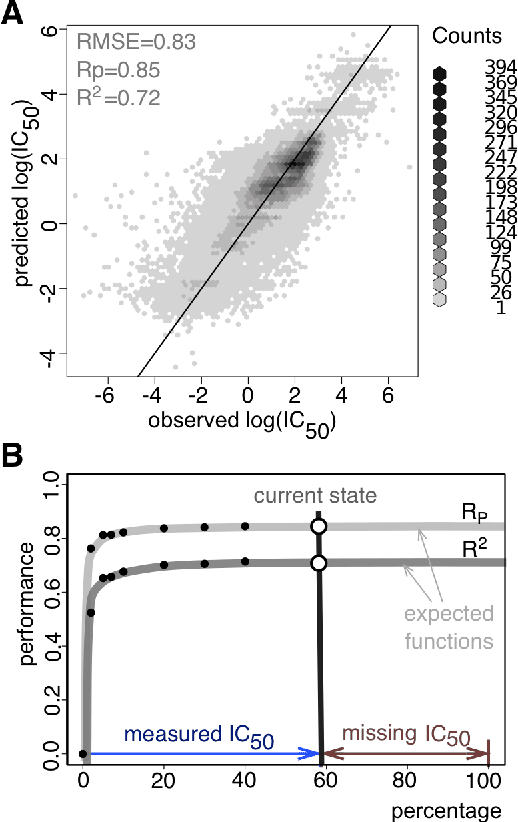
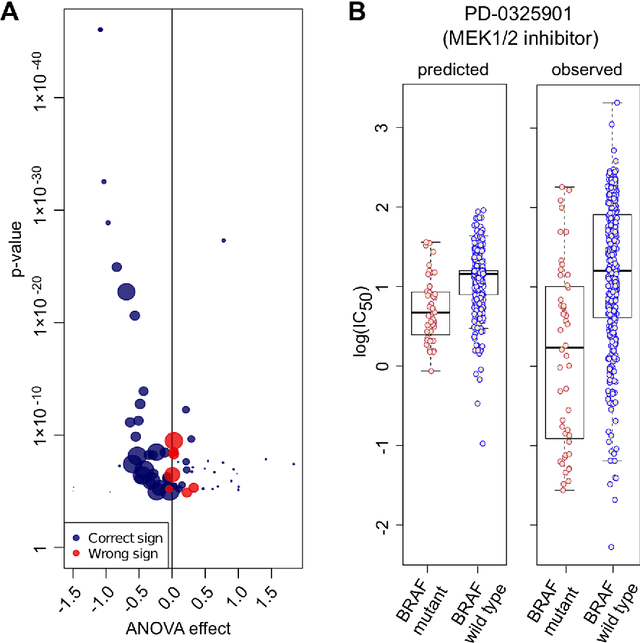
Abstract:Predicting the response of a specific cancer to a therapy is a major goal in modern oncology that should ultimately lead to a personalised treatment. High-throughput screenings of potentially active compounds against a panel of genomically heterogeneous cancer cell lines have unveiled multiple relationships between genomic alterations and drug responses. Various computational approaches have been proposed to predict sensitivity based on genomic features, while others have used the chemical properties of the drugs to ascertain their effect. In an effort to integrate these complementary approaches, we developed machine learning models to predict the response of cancer cell lines to drug treatment, quantified through IC50 values, based on both the genomic features of the cell lines and the chemical properties of the considered drugs. Models predicted IC50 values in a 8-fold cross-validation and an independent blind test with coefficient of determination R2 of 0.72 and 0.64 respectively. Furthermore, models were able to predict with comparable accuracy (R2 of 0.61) IC50s of cell lines from a tissue not used in the training stage. Our in silico models can be used to optimise the experimental design of drug-cell screenings by estimating a large proportion of missing IC50 values rather than experimentally measure them. The implications of our results go beyond virtual drug screening design: potentially thousands of drugs could be probed in silico to systematically test their potential efficacy as anti-tumour agents based on their structure, thus providing a computational framework to identify new drug repositioning opportunities as well as ultimately be useful for personalized medicine by linking the genomic traits of patients to drug sensitivity.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge