Ewan Cameron
Variational Learning on Aggregate Outputs with Gaussian Processes
May 22, 2018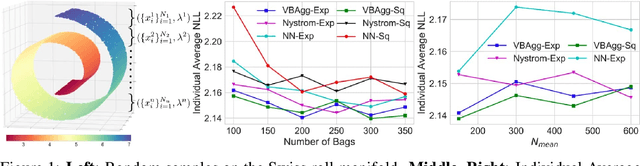
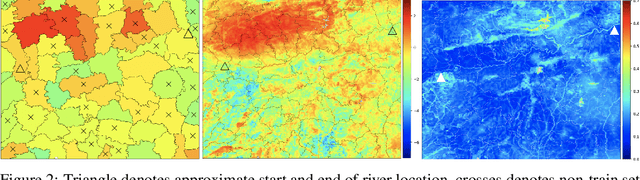
Abstract:While a typical supervised learning framework assumes that the inputs and the outputs are measured at the same levels of granularity, many applications, including global mapping of disease, only have access to outputs at a much coarser level than that of the inputs. Aggregation of outputs makes generalization to new inputs much more difficult. We consider an approach to this problem based on variational learning with a model of output aggregation and Gaussian processes, where aggregation leads to intractability of the standard evidence lower bounds. We propose new bounds and tractable approximations, leading to improved prediction accuracy and scalability to large datasets, while explicitly taking uncertainty into account. We develop a framework which extends to several types of likelihoods, including the Poisson model for aggregated count data. We apply our framework to a challenging and important problem, the fine-scale spatial modelling of malaria incidence, with over 1 million observations.
Improved prediction accuracy for disease risk mapping using Gaussian Process stacked generalisation
Dec 10, 2016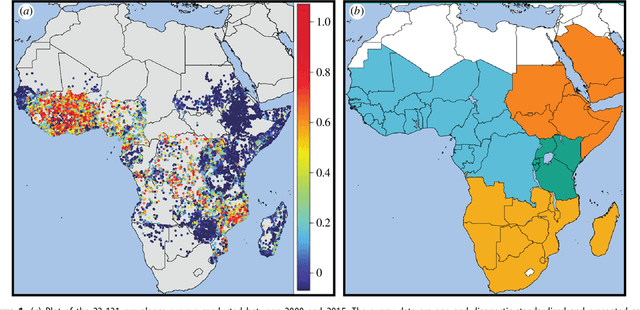
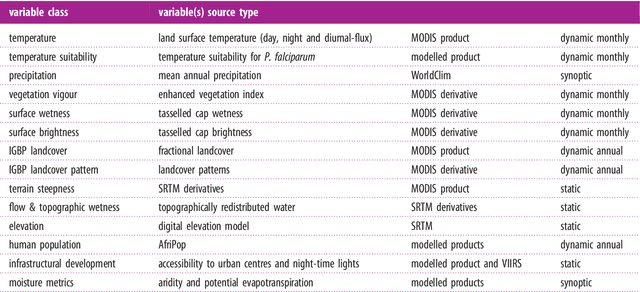
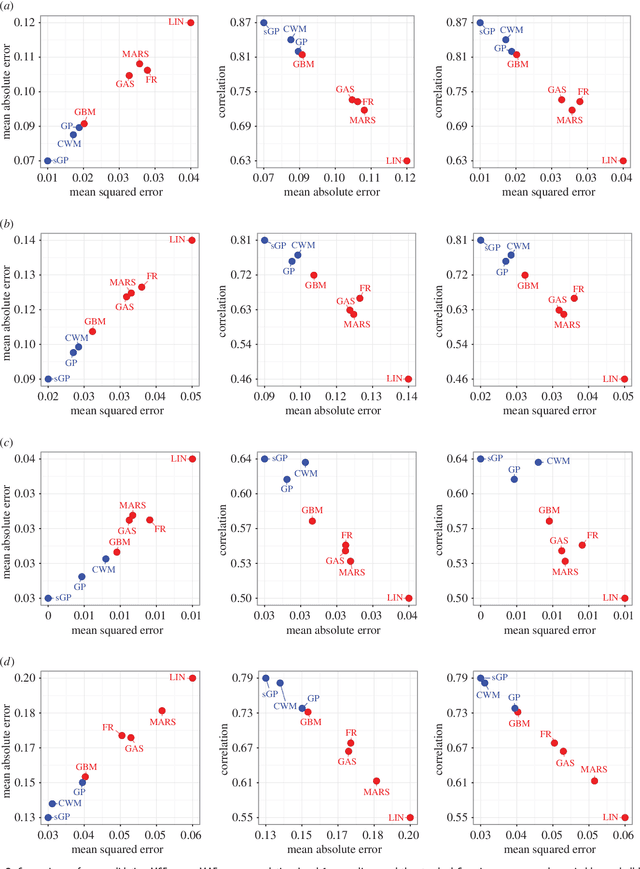
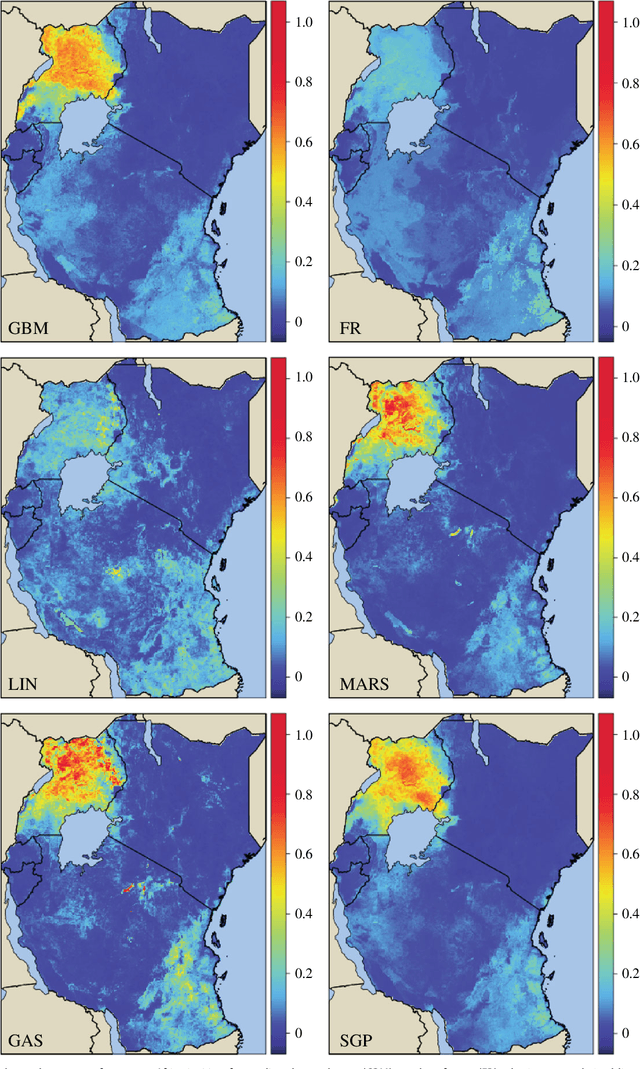
Abstract:Maps of infectious disease---charting spatial variations in the force of infection, degree of endemicity, and the burden on human health---provide an essential evidence base to support planning towards global health targets. Contemporary disease mapping efforts have embraced statistical modelling approaches to properly acknowledge uncertainties in both the available measurements and their spatial interpolation. The most common such approach is that of Gaussian process regression, a mathematical framework comprised of two components: a mean function harnessing the predictive power of multiple independent variables, and a covariance function yielding spatio-temporal shrinkage against residual variation from the mean. Though many techniques have been developed to improve the flexibility and fitting of the covariance function, models for the mean function have typically been restricted to simple linear terms. For infectious diseases, known to be driven by complex interactions between environmental and socio-economic factors, improved modelling of the mean function can greatly boost predictive power. Here we present an ensemble approach based on stacked generalisation that allows for multiple, non-linear algorithmic mean functions to be jointly embedded within the Gaussian process framework. We apply this method to mapping Plasmodium falciparum prevalence data in Sub-Saharan Africa and show that the generalised ensemble approach markedly out-performs any individual method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge