Enze Xu
Fourier-enhanced Neural Networks For Systems Biology Applications
Feb 10, 2025Abstract:In the field of systems biology, differential equations are commonly used to model biological systems, but solving them for large-scale and complex systems can be computationally expensive. Recently, the integration of machine learning and mathematical modeling has offered new opportunities for scientific discoveries in biology and health. The emerging physics-informed neural network (PINN) has been proposed as a solution to this problem. However, PINN can be computationally expensive and unreliable for complex biological systems. To address these issues, we propose the Fourier-enhanced Neural Networks for systems biology (SB-FNN). SB-FNN uses an embedded Fourier neural network with an adaptive activation function and a cyclic penalty function to optimize the prediction of biological dynamics, particularly for biological systems that exhibit oscillatory patterns. Experimental results demonstrate that SB-FNN achieves better performance and is more efficient than PINN for handling complex biological models. Experimental results on cellular and population models demonstrate that SB-FNN outperforms PINN in both accuracy and efficiency, making it a promising alternative approach for handling complex biological models. The proposed method achieved better performance on six biological models and is expected to replace PINN as the most advanced method in systems biology.
Neural Symbolic Regression using Control Variables
Jun 07, 2023Abstract:Symbolic regression (SR) is a powerful technique for discovering the analytical mathematical expression from data, finding various applications in natural sciences due to its good interpretability of results. However, existing methods face scalability issues when dealing with complex equations involving multiple variables. To address this challenge, we propose SRCV, a novel neural symbolic regression method that leverages control variables to enhance both accuracy and scalability. The core idea is to decompose multi-variable symbolic regression into a set of single-variable SR problems, which are then combined in a bottom-up manner. The proposed method involves a four-step process. First, we learn a data generator from observed data using deep neural networks (DNNs). Second, the data generator is used to generate samples for a certain variable by controlling the input variables. Thirdly, single-variable symbolic regression is applied to estimate the corresponding mathematical expression. Lastly, we repeat steps 2 and 3 by gradually adding variables one by one until completion. We evaluate the performance of our method on multiple benchmark datasets. Experimental results demonstrate that the proposed SRCV significantly outperforms state-of-the-art baselines in discovering mathematical expressions with multiple variables. Moreover, it can substantially reduce the search space for symbolic regression. The source code will be made publicly available upon publication.
Pathology Steered Stratification Network for Subtype Identification in Alzheimer's Disease
Oct 12, 2022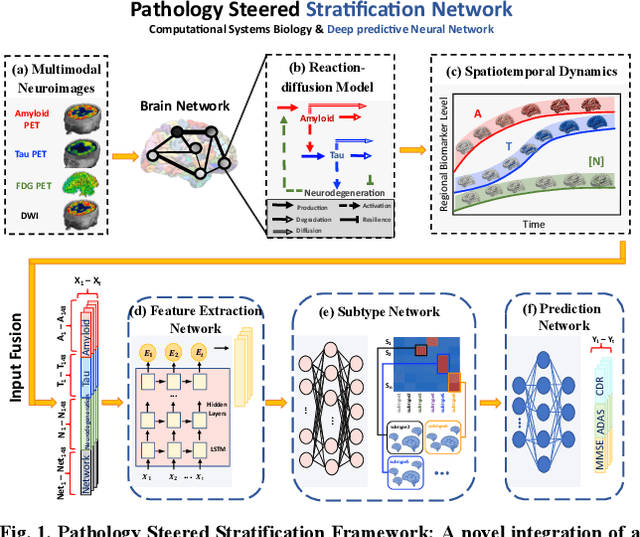
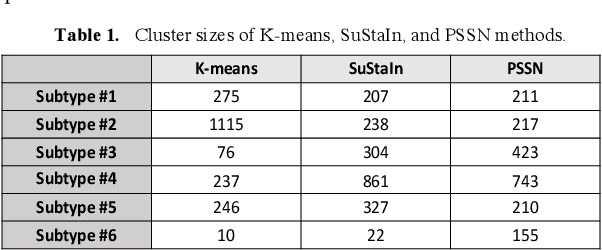
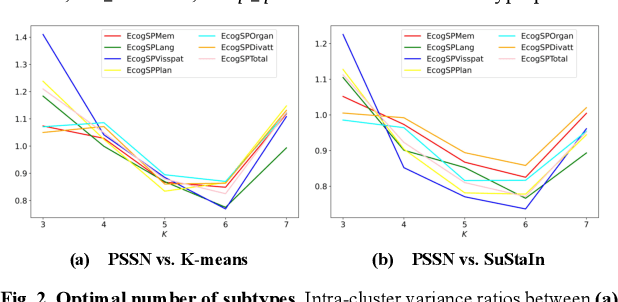
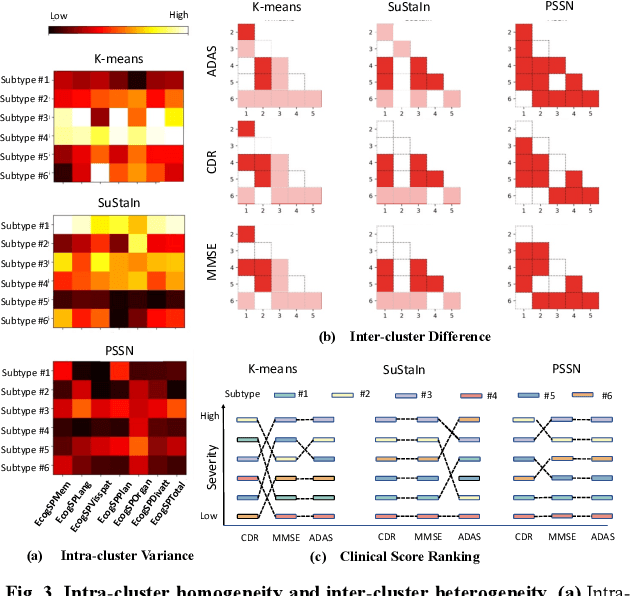
Abstract:Alzheimer's disease (AD) is a heterogeneous, multifactorial neurodegenerative disorder characterized by beta-amyloid, pathologic tau, and neurodegeneration. The massive heterogeneity between neurobiological examinations and clinical assessment is the current biggest challenge in the early diagnosis of Alzheimer's disease, urging for a comprehensive stratification of the aging population that is defined by reliable neurobiological biomarkers and closely associated with clinical outcomes. However, existing statistical inference approaches in neuroimaging studies of AD subtype identification fail to take into account the neuropathological domain knowledge, which could lead to ill-posed results that are sometimes inconsistent with neurological principles. To fill this knowledge gap, we propose a novel pathology steered stratification network (PSSN) that integrates mainstream AD pathology with multimodal longitudinal neuroimaging data to categorize the aging population. By combining theory-based biological modeling and data-driven deep learning, this cross-disciplinary approach can not only generate long-term biomarker prediction consistent with the end-state of individuals but also stratifies subjects into fine-grained subtypes with distinct neurological underpinnings, where ag-ing brains within the same subtype share com-mon biological behaviors that emerge as similar trajectories of cognitive decline. Our stratification outperforms K-means and SuStaIn in both inter-cluster heterogeneity and intra-cluster homogeneity of various clinical scores. Importantly, we identify six subtypes spanning AD spectrum, where each subtype exhibits a distinctive biomarker pattern that is consistent with its clinical outcome. A disease evolutionary graph is further provided by quantifying subtype transition probabilities, which may assist pre-symptomatic diagnosis and guide therapeutic treatments.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge