Emmanuel Iarussi
Recursive Variational Autoencoders for 3D Blood Vessel Generative Modeling
Jun 17, 2025Abstract:Anatomical trees play an important role in clinical diagnosis and treatment planning. Yet, accurately representing these structures poses significant challenges owing to their intricate and varied topology and geometry. Most existing methods to synthesize vasculature are rule based, and despite providing some degree of control and variation in the structures produced, they fail to capture the diversity and complexity of actual anatomical data. We developed a Recursive variational Neural Network (RvNN) that fully exploits the hierarchical organization of the vessel and learns a low-dimensional manifold encoding branch connectivity along with geometry features describing the target surface. After training, the RvNN latent space can be sampled to generate new vessel geometries. By leveraging the power of generative neural networks, we generate 3D models of blood vessels that are both accurate and diverse, which is crucial for medical and surgical training, hemodynamic simulations, and many other purposes. These results closely resemble real data, achieving high similarity in vessel radii, length, and tortuosity across various datasets, including those with aneurysms. To the best of our knowledge, this work is the first to utilize this technique for synthesizing blood vessels.
VesselGPT: Autoregressive Modeling of Vascular Geometry
May 19, 2025Abstract:Anatomical trees are critical for clinical diagnosis and treatment planning, yet their complex and diverse geometry make accurate representation a significant challenge. Motivated by the latest advances in large language models, we introduce an autoregressive method for synthesizing anatomical trees. Our approach first embeds vessel structures into a learned discrete vocabulary using a VQ-VAE architecture, then models their generation autoregressively with a GPT-2 model. This method effectively captures intricate geometries and branching patterns, enabling realistic vascular tree synthesis. Comprehensive qualitative and quantitative evaluations reveal that our technique achieves high-fidelity tree reconstruction with compact discrete representations. Moreover, our B-spline representation of vessel cross-sections preserves critical morphological details that are often overlooked in previous' methods parameterizations. To the best of our knowledge, this work is the first to generate blood vessels in an autoregressive manner. Code, data, and trained models will be made available.
DUDF: Differentiable Unsigned Distance Fields with Hyperbolic Scaling
Feb 14, 2024Abstract:In recent years, there has been a growing interest in training Neural Networks to approximate Unsigned Distance Fields (UDFs) for representing open surfaces in the context of 3D reconstruction. However, UDFs are non-differentiable at the zero level set which leads to significant errors in distances and gradients, generally resulting in fragmented and discontinuous surfaces. In this paper, we propose to learn a hyperbolic scaling of the unsigned distance field, which defines a new Eikonal problem with distinct boundary conditions. This allows our formulation to integrate seamlessly with state-of-the-art continuously differentiable implicit neural representation networks, largely applied in the literature to represent signed distance fields. Our approach not only addresses the challenge of open surface representation but also demonstrates significant improvement in reconstruction quality and training performance. Moreover, the unlocked field's differentiability allows the accurate computation of essential topological properties such as normal directions and curvatures, pervasive in downstream tasks such as rendering. Through extensive experiments, we validate our approach across various data sets and against competitive baselines. The results demonstrate enhanced accuracy and up to an order of magnitude increase in speed compared to previous methods.
VesselVAE: Recursive Variational Autoencoders for 3D Blood Vessel Synthesis
Jul 07, 2023Abstract:We present a data-driven generative framework for synthesizing blood vessel 3D geometry. This is a challenging task due to the complexity of vascular systems, which are highly variating in shape, size, and structure. Existing model-based methods provide some degree of control and variation in the structures produced, but fail to capture the diversity of actual anatomical data. We developed VesselVAE, a recursive variational Neural Network that fully exploits the hierarchical organization of the vessel and learns a low-dimensional manifold encoding branch connectivity along with geometry features describing the target surface. After training, the VesselVAE latent space can be sampled to generate new vessel geometries. To the best of our knowledge, this work is the first to utilize this technique for synthesizing blood vessels. We achieve similarities of synthetic and real data for radius (.97), length (.95), and tortuosity (.96). By leveraging the power of deep neural networks, we generate 3D models of blood vessels that are both accurate and diverse, which is crucial for medical and surgical training, hemodynamic simulations, and many other purposes.
Learning normal asymmetry representations for homologous brain structures
Jun 27, 2023Abstract:Although normal homologous brain structures are approximately symmetrical by definition, they also have shape differences due to e.g. natural ageing. On the other hand, neurodegenerative conditions induce their own changes in this asymmetry, making them more pronounced or altering their location. Identifying when these alterations are due to a pathological deterioration is still challenging. Current clinical tools rely either on subjective evaluations, basic volume measurements or disease-specific deep learning models. This paper introduces a novel method to learn normal asymmetry patterns in homologous brain structures based on anomaly detection and representation learning. Our framework uses a Siamese architecture to map 3D segmentations of left and right hemispherical sides of a brain structure to a normal asymmetry embedding space, learned using a support vector data description objective. Being trained using healthy samples only, it can quantify deviations-from-normal-asymmetry patterns in unseen samples by measuring the distance of their embeddings to the center of the learned normal space. We demonstrate in public and in-house sets that our method can accurately characterize normal asymmetries and detect pathological alterations due to Alzheimer's disease and hippocampal sclerosis, even though no diseased cases were accessed for training. Our source code is available at https://github.com/duiliod/DeepNORHA.
Generative Modelling of 3D in-silico Spongiosa with Controllable Micro-Structural Parameters
Sep 23, 2020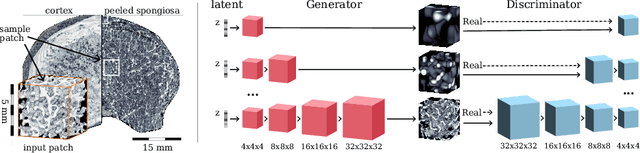
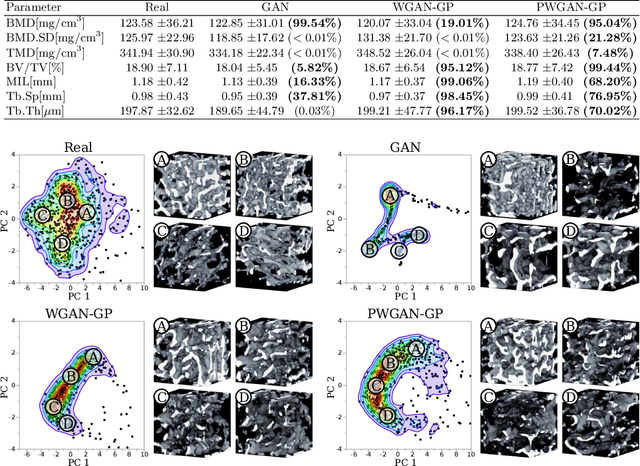
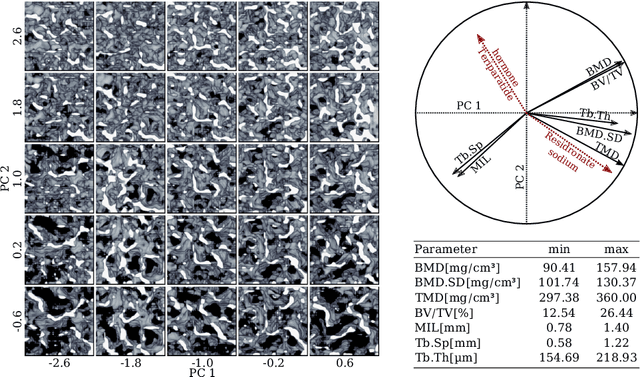
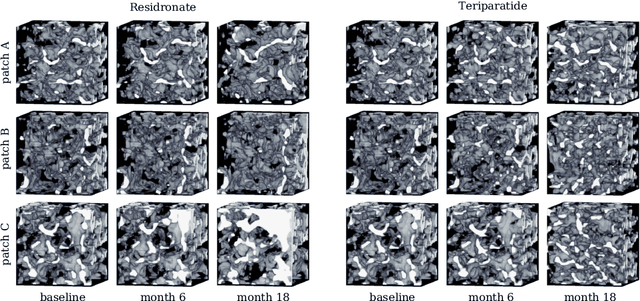
Abstract:Research in vertebral bone micro-structure generally requires costly procedures to obtain physical scans of real bone with a specific pathology under study, since no methods are available yet to generate realistic bone structures in-silico. Here we propose to apply recent advances in generative adversarial networks (GANs) to develop such a method. We adapted style-transfer techniques, which have been largely used in other contexts, in order to transfer style between image pairs while preserving its informational content. In a first step, we trained a volumetric generative model in a progressive manner using a Wasserstein objective and gradient penalty (PWGAN-GP) to create patches of realistic bone structure in-silico. The training set contained 7660 purely spongeous bone samples from twelve human vertebrae (T12 or L1) with isotropic resolution of 164um and scanned with a high resolution peripheral quantitative CT (Scanco XCT). After training, we generated new samples with tailored micro-structure properties by optimizing a vector z in the learned latent space. To solve this optimization problem, we formulated a differentiable goal function that leads to valid samples while compromising the appearance (content) with target 3D properties (style). Properties of the learned latent space effectively matched the data distribution. Furthermore, we were able to simulate the resulting bone structure after deterioration or treatment effects of osteoporosis therapies based only on expected changes of micro-structural parameters. Our method allows to generate a virtually infinite number of patches of realistic bone micro-structure, and thereby likely serves for the development of bone-biomarkers and to simulate bone therapies in advance.
SketchZooms: Deep multi-view descriptors for matching line drawings
Nov 29, 2019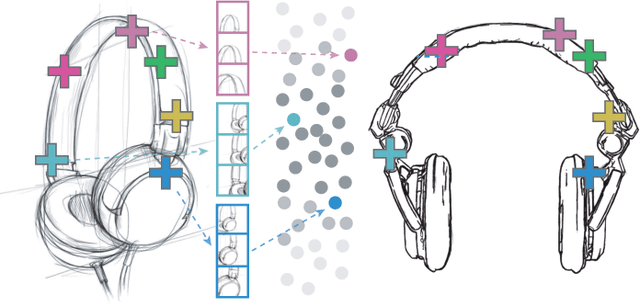
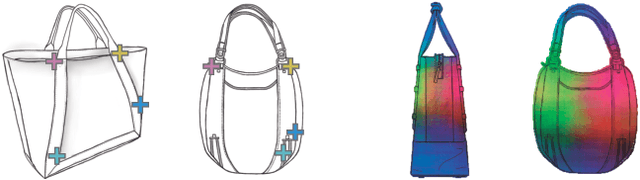
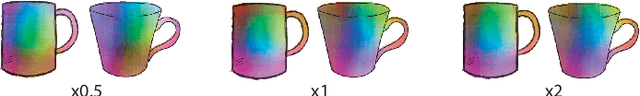
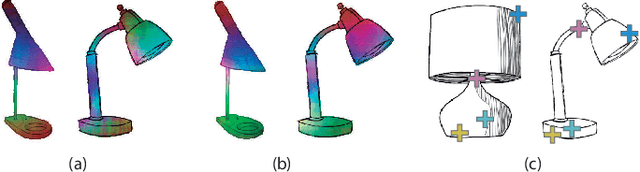
Abstract:Finding point-wise correspondences between images is a long-standing problem in computer vision. Corresponding sketch images is particularly challenging due to the varying nature of human style, projection distortions and viewport changes. In this paper we present a feature descriptor targeting line drawings learned from a 3D shape data set. Our descriptors are designed to locally match image pairs where the object of interest belongs to the same semantic category, yet still differ drastically in shape and projection angle. We build our descriptors by means of a Convolutional Neural Network (CNN) trained in a triplet fashion. The goal is to embed semantically similar anchor points close to one another, and to pull the embeddings of different points far apart. To learn the descriptors space, the network is fed with a succession of zoomed views from the input sketches. We have specifically crafted a data set of synthetic sketches using a non-photorealistic rendering algorithm over a large collection of part-based registered 3D models. Once trained, our network can generate descriptors for every pixel in an input image. Furthermore, our network is able to generalize well to unseen sketches hand-drawn by humans, outperforming state-of-the-art descriptors on the evaluated matching tasks. Our descriptors can be used to obtain sparse and dense correspondences between image pairs. We evaluate our method against a baseline of correspondences data collected from expert designers, in addition to comparisons with descriptors that have been proven effective in sketches. Finally, we demonstrate applications showing the usefulness of our multi-view descriptors.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge