Elazer R. Edelman
The Dissipation Theory of Aging: A Quantitative Analysis Using a Cellular Aging Map
Apr 17, 2025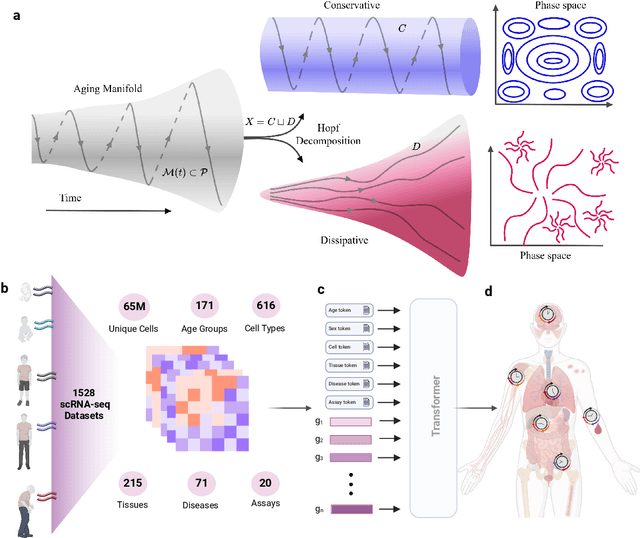
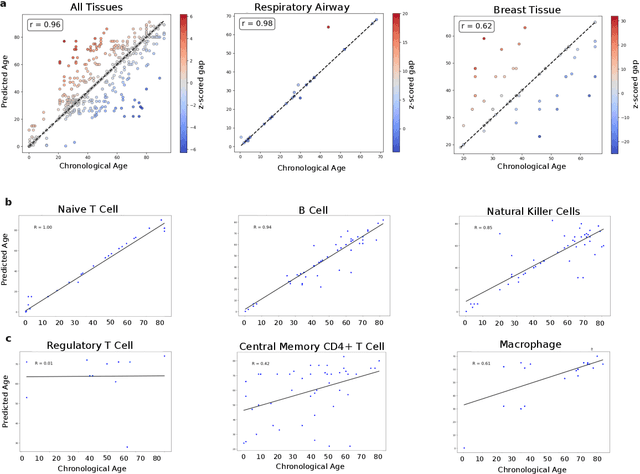
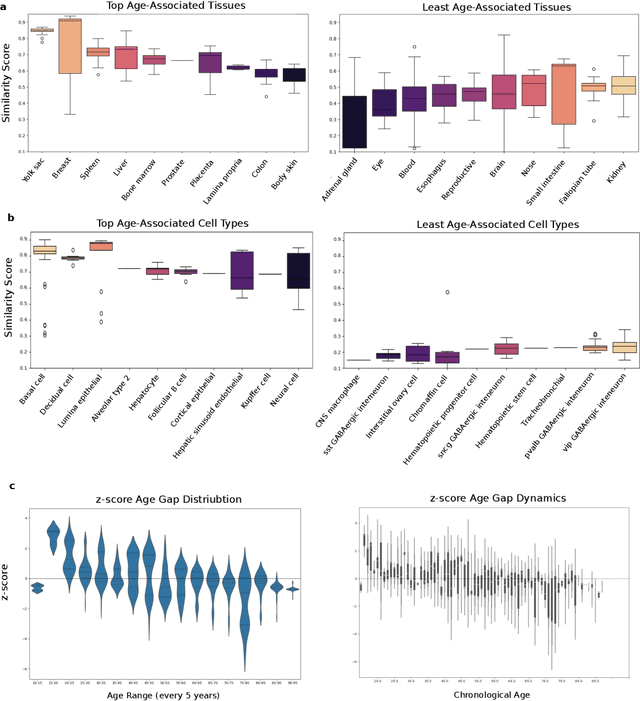
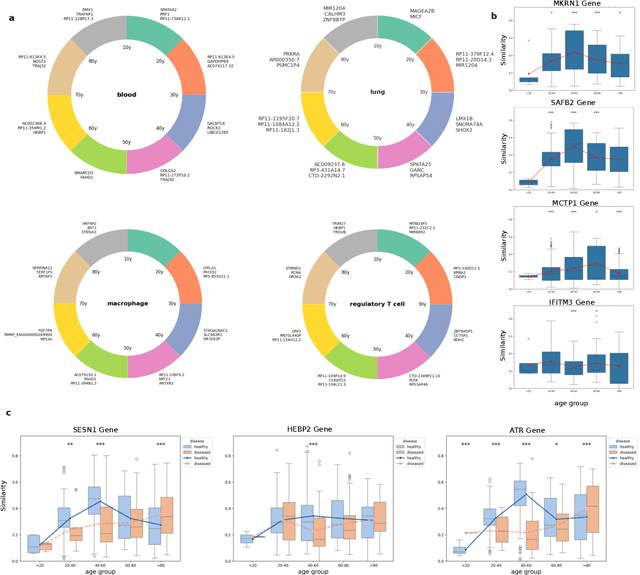
Abstract:We propose a new theory for aging based on dynamical systems and provide a data-driven computational method to quantify the changes at the cellular level. We use ergodic theory to decompose the dynamics of changes during aging and show that aging is fundamentally a dissipative process within biological systems, akin to dynamical systems where dissipation occurs due to non-conservative forces. To quantify the dissipation dynamics, we employ a transformer-based machine learning algorithm to analyze gene expression data, incorporating age as a token to assess how age-related dissipation is reflected in the embedding space. By evaluating the dynamics of gene and age embeddings, we provide a cellular aging map (CAM) and identify patterns indicative of divergence in gene embedding space, nonlinear transitions, and entropy variations during aging for various tissues and cell types. Our results provide a novel perspective on aging as a dissipative process and introduce a computational framework that enables measuring age-related changes with molecular resolution.
Cellular Development Follows the Path of Minimum Action
Apr 10, 2025Abstract:Cellular development follows a stochastic yet rule-governed trajectory, though the underlying principles remain elusive. Here, we propose that cellular development follows paths of least action, aligning with foundational physical laws that govern dynamic systems across nature. We introduce a computational framework that takes advantage of the deep connection between the principle of least action and maximum entropy to model developmental processes using Transformers architecture. This approach enables precise quantification of entropy production, information flow curvature, and local irreversibility for developmental asymmetry in single-cell RNA sequence data. Within this unified framework, we provide interpretable metrics: entropy to capture exploration-exploitation trade-offs, curvature to assess plasticity-elasticity dynamics, and entropy production to characterize dedifferentiation and transdifferentiation. We validate our method across both single-cell and embryonic development datasets, demonstrating its ability to reveal hidden thermodynamic and informational constraints shaping cellular fate decisions.
A Diffusion Model for Simulation Ready Coronary Anatomy with Morpho-skeletal Control
Jul 23, 2024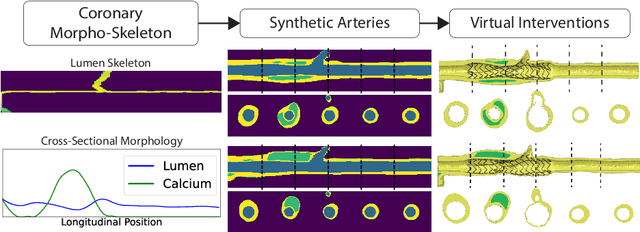
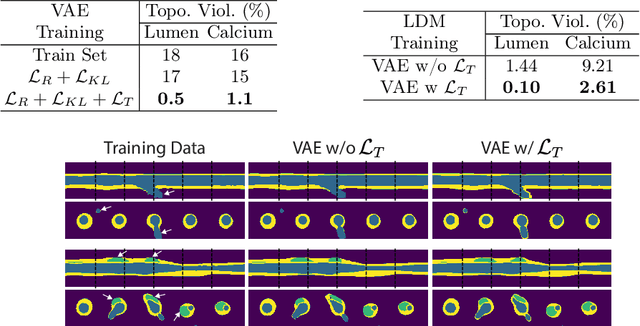
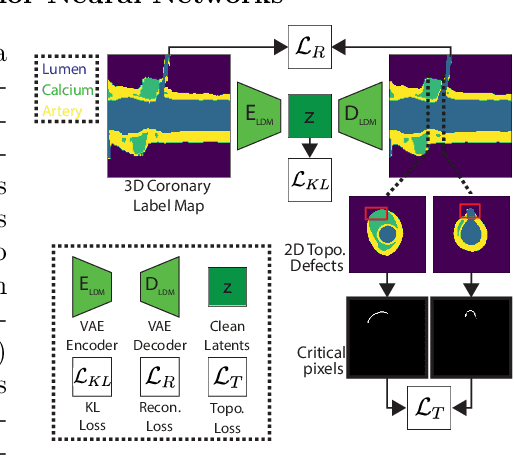
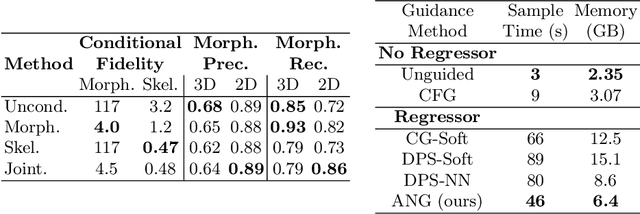
Abstract:Virtual interventions enable the physics-based simulation of device deployment within coronary arteries. This framework allows for counterfactual reasoning by deploying the same device in different arterial anatomies. However, current methods to create such counterfactual arteries face a trade-off between controllability and realism. In this study, we investigate how Latent Diffusion Models (LDMs) can custom synthesize coronary anatomy for virtual intervention studies based on mid-level anatomic constraints such as topological validity, local morphological shape, and global skeletal structure. We also extend diffusion model guidance strategies to the context of morpho-skeletal conditioning and propose a novel guidance method for continuous attributes that adaptively updates the negative guiding condition throughout sampling. Our framework enables the generation and editing of coronary anatomy in a controllable manner, allowing device designers to derive mechanistic insights regarding anatomic variation and simulated device deployment.
Probing the Limits and Capabilities of Diffusion Models for the Anatomic Editing of Digital Twins
Dec 30, 2023Abstract:Numerical simulations can model the physical processes that govern cardiovascular device deployment. When such simulations incorporate digital twins; computational models of patient-specific anatomy, they can expedite and de-risk the device design process. Nonetheless, the exclusive use of patient-specific data constrains the anatomic variability which can be precisely or fully explored. In this study, we investigate the capacity of Latent Diffusion Models (LDMs) to edit digital twins to create anatomic variants, which we term digital siblings. Digital twins and their corresponding siblings can serve as the basis for comparative simulations, enabling the study of how subtle anatomic variations impact the simulated deployment of cardiovascular devices, as well as the augmentation of virtual cohorts for device assessment. However, while diffusion models have been characterized in their ability to edit natural images, their capacity to anatomically edit digital twins has yet to be studied. Using a case example centered on 3D digital twins of cardiac anatomy, we implement various methods for generating digital siblings and characterize them through morphological and topological analyses. We specifically edit digital twins to introduce anatomic variation at different spatial scales and within localized regions, demonstrating the existence of bias towards common anatomic features. We further show that such anatomic bias can be leveraged for virtual cohort augmentation through selective editing, partially alleviating issues related to dataset imbalance and lack of diversity. Our experimental framework thus delineates the limits and capabilities of using latent diffusion models in synthesizing anatomic variation for in silico trials.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge