Dorota Toczydlowska
BioNeMo Framework: a modular, high-performance library for AI model development in drug discovery
Nov 15, 2024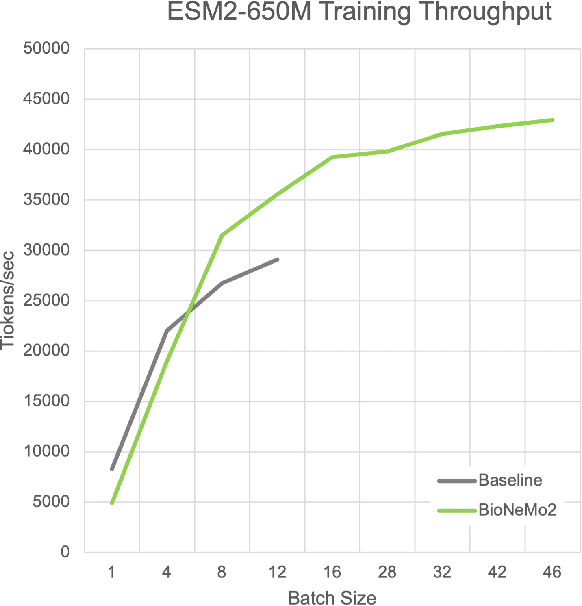
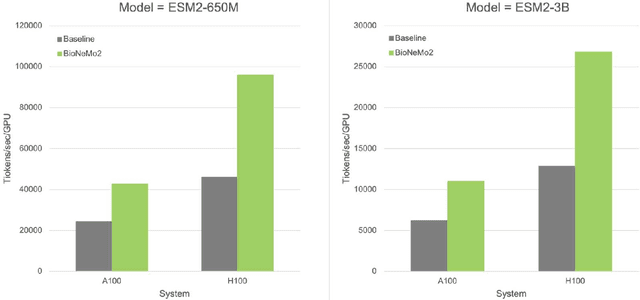
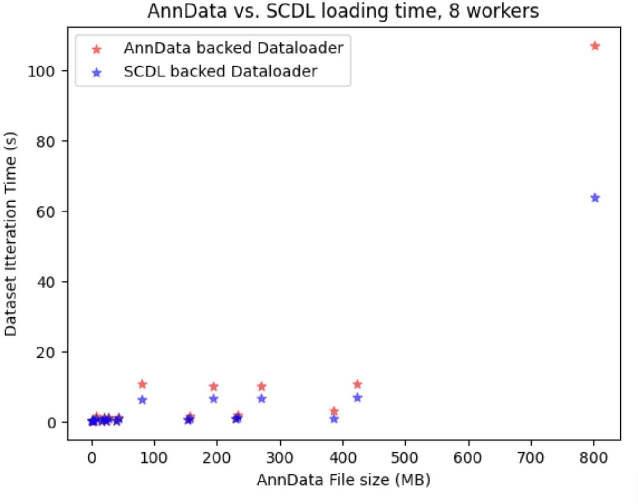
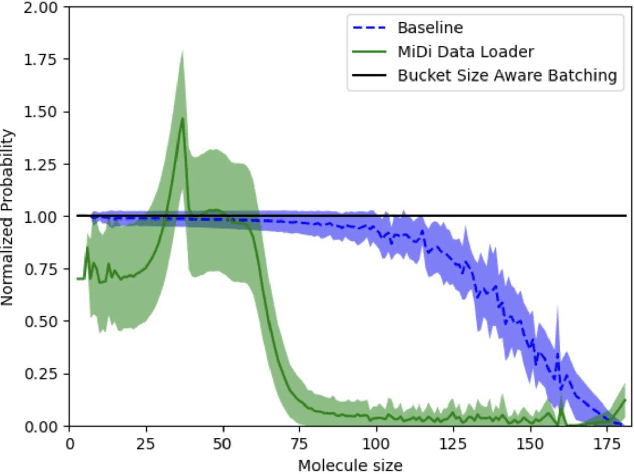
Abstract:Artificial Intelligence models encoding biology and chemistry are opening new routes to high-throughput and high-quality in-silico drug development. However, their training increasingly relies on computational scale, with recent protein language models (pLM) training on hundreds of graphical processing units (GPUs). We introduce the BioNeMo Framework to facilitate the training of computational biology and chemistry AI models across hundreds of GPUs. Its modular design allows the integration of individual components, such as data loaders, into existing workflows and is open to community contributions. We detail technical features of the BioNeMo Framework through use cases such as pLM pre-training and fine-tuning. On 256 NVIDIA A100s, BioNeMo Framework trains a three billion parameter BERT-based pLM on over one trillion tokens in 4.2 days. The BioNeMo Framework is open-source and free for everyone to use.
Parsimonious Feature Extraction Methods: Extending Robust Probabilistic Projections with Generalized Skew-t
Sep 24, 2020
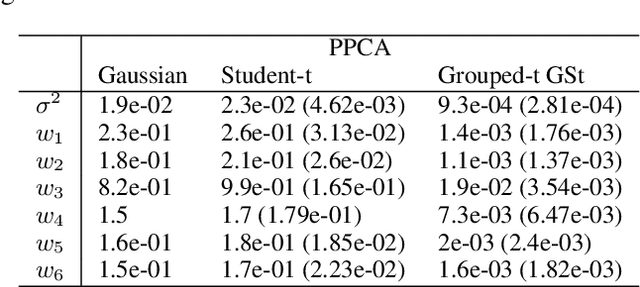
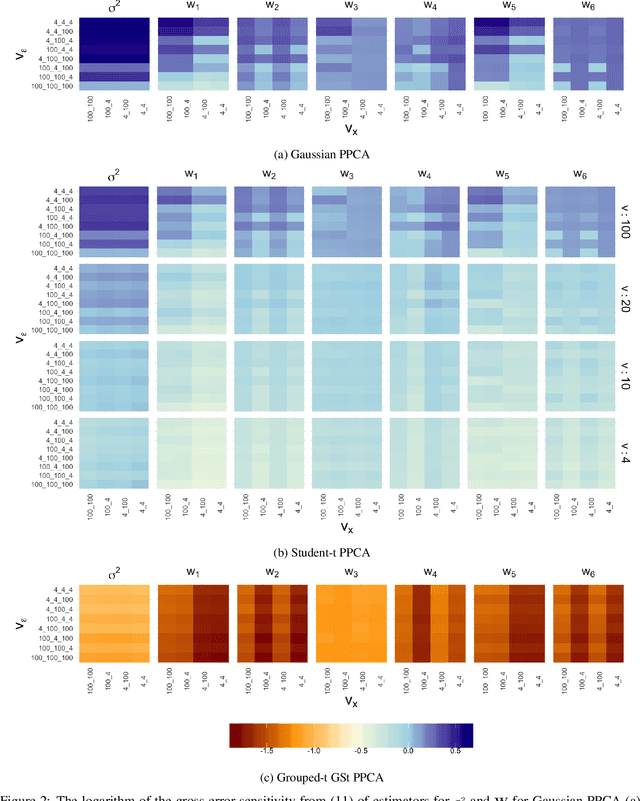
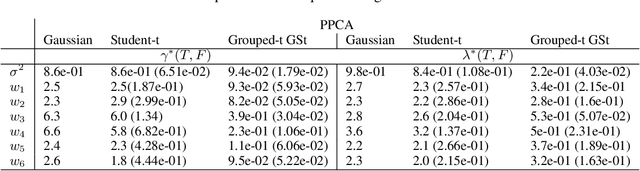
Abstract:We propose a novel generalisation to the Student-t Probabilistic Principal Component methodology which: (1) accounts for an asymmetric distribution of the observation data; (2) is a framework for grouped and generalised multiple-degree-of-freedom structures, which provides a more flexible approach to modelling groups of marginal tail dependence in the observation data; and (3) separates the tail effect of the error terms and factors. The new feature extraction methods are derived in an incomplete data setting to efficiently handle the presence of missing values in the observation vector. We discuss various special cases of the algorithm being a result of simplified assumptions on the process generating the data. The applicability of the new framework is illustrated on a data set that consists of crypto currencies with the highest market capitalisation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge