Dominik J. E. Waibel
A Diffusion Model Predicts 3D Shapes from 2D Microscopy Images
Aug 31, 2022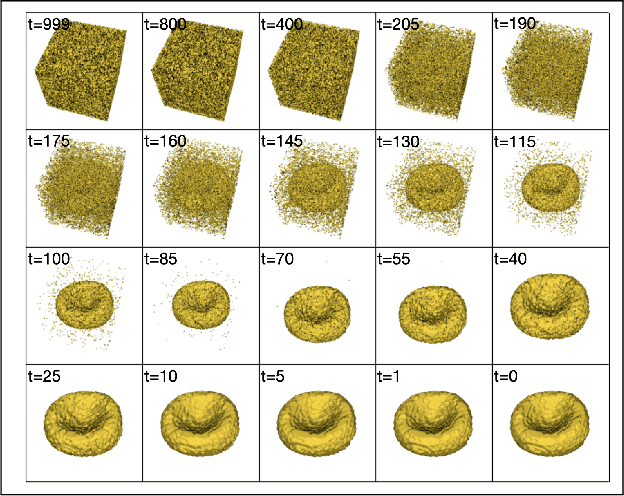
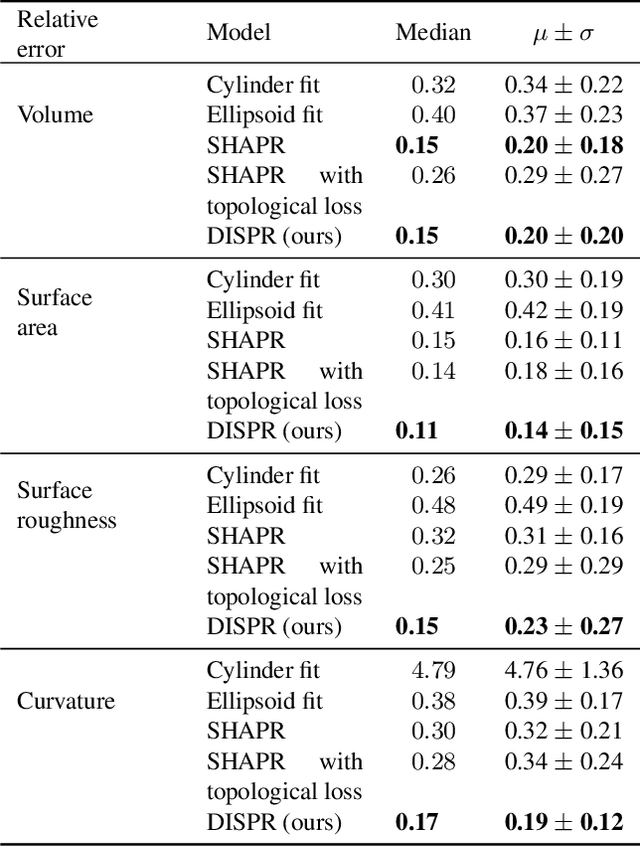
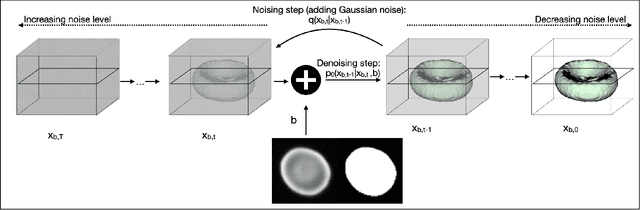
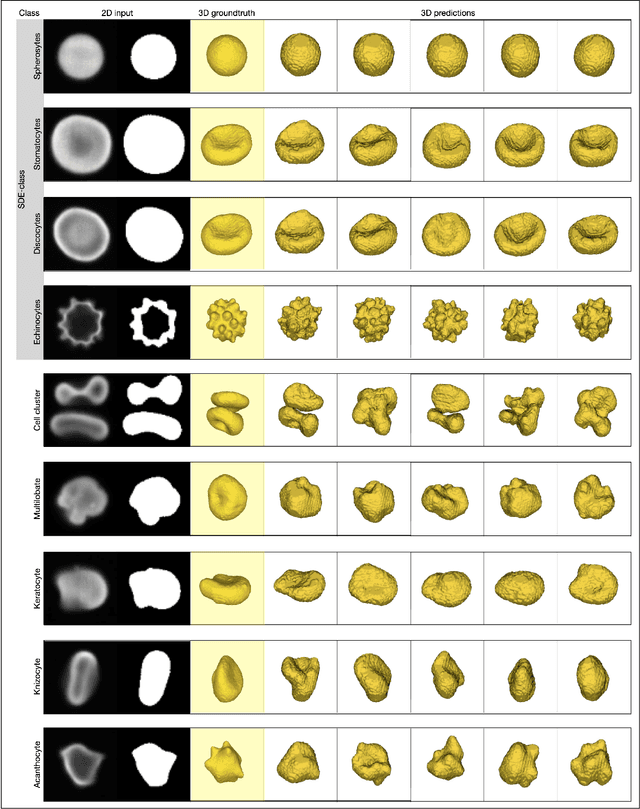
Abstract:Diffusion models are a class of generative models, showing superior performance as compared to other generative models in creating realistic images when trained on natural image datasets. We introduce DISPR, a diffusion-based model for solving the inverse problem of three-dimensional (3D) cell shape prediction from two-dimensional (2D) single cell microscopy images. Using the 2D microscopy image as a prior, DISPR is conditioned to predict realistic 3D shape reconstructions. To showcase the applicability of DISPR as a data augmentation tool in a feature-based single cell classification task, we extract morphological features from the cells grouped into six highly imbalanced classes. Adding features from predictions of DISPR to the three minority classes improved the macro F1 score from $F1_\text{macro} = 55.2 \pm 4.6\%$ to $F1_\text{macro} = 72.2 \pm 4.9\%$. With our method being the first to employ a diffusion-based model in this context, we demonstrate that diffusion models can be applied to inverse problems in 3D, and that they learn to reconstruct 3D shapes with realistic morphological features from 2D microscopy images.
Capturing Shape Information with Multi-Scale Topological Loss Terms for 3D Reconstruction
Mar 03, 2022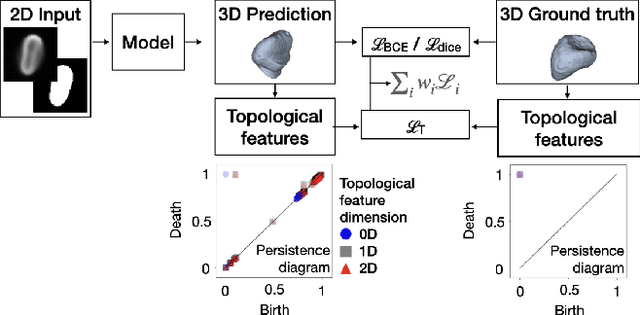
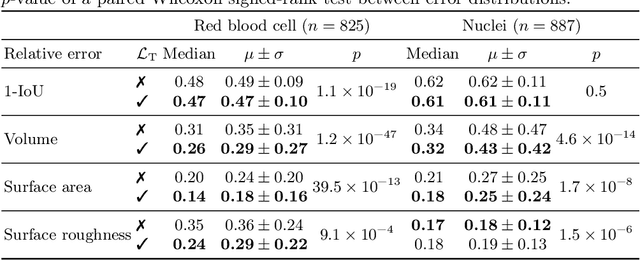
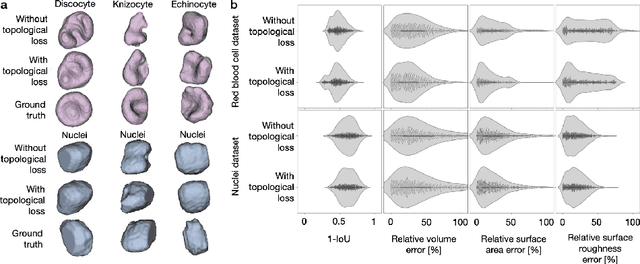
Abstract:Reconstructing 3D objects from 2D images is both challenging for our brains and machine learning algorithms. To support this spatial reasoning task, contextual information about the overall shape of an object is critical. However, such information is not captured by established loss terms (e.g. Dice loss). We propose to complement geometrical shape information by including multi-scale topological features, such as connected components, cycles, and voids, in the reconstruction loss. Our method calculates topological features from 3D volumetric data based on cubical complexes and uses an optimal transport distance to guide the reconstruction process. This topology-aware loss is fully differentiable, computationally efficient, and can be added to any neural network. We demonstrate the utility of our loss by incorporating it into SHAPR, a model for predicting the 3D cell shape of individual cells based on 2D microscopy images. Using a hybrid loss that leverages both geometrical and topological information of single objects to assess their shape, we find that topological information substantially improves the quality of reconstructions, thus highlighting its ability to extract more relevant features from image datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge