Domenico L. Gatti
Molecular dynamics without molecules: searching the conformational space of proteins with generative neural networks
Jun 09, 2022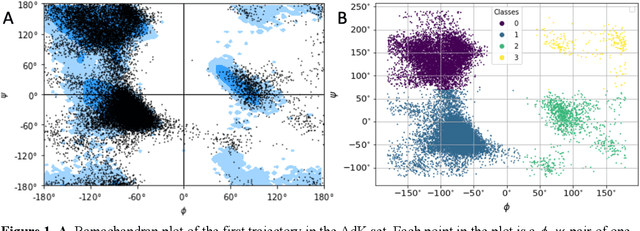
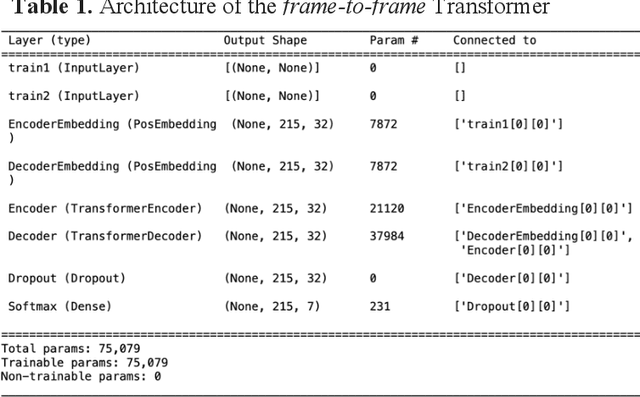

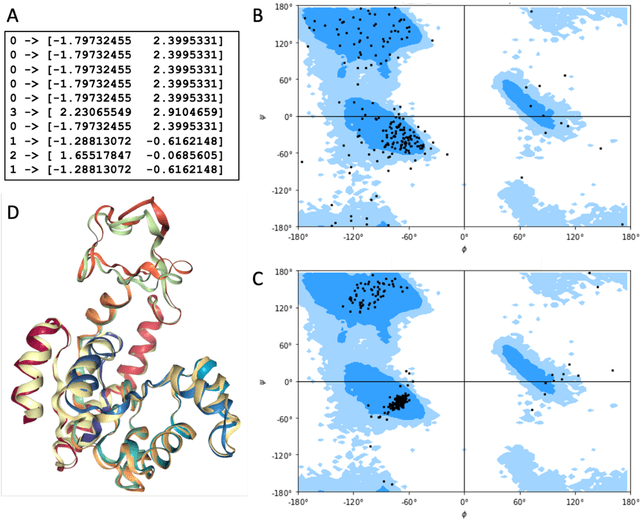
Abstract:All-atom and coarse-grained molecular dynamics are two widely used computational tools to study the conformational states of proteins. Yet, these two simulation methods suffer from the fact that without access to supercomputing resources, the time and length scales at which these states become detectable are difficult to achieve. One alternative to such methods is based on encoding the atomistic trajectory of molecular dynamics as a shorthand version devoid of physical particles, and then learning to propagate the encoded trajectory through the use of artificial intelligence. Here we show that a simple textual representation of the frames of molecular dynamics trajectories as vectors of Ramachandran basin classes retains most of the structural information of the full atomistic representation of a protein in each frame, and can be used to generate equivalent atom-less trajectories suitable to train different types of generative neural networks. In turn, the trained generative models can be used to extend indefinitely the atom-less dynamics or to sample the conformational space of proteins from their representation in the models latent space. We define intuitively this methodology as molecular dynamics without molecules, and show that it enables to cover physically relevant states of proteins that are difficult to access with traditional molecular dynamics.
CXR-Net: An Artificial Intelligence Pipeline for Quick Covid-19 Screening of Chest X-Rays
Feb 26, 2021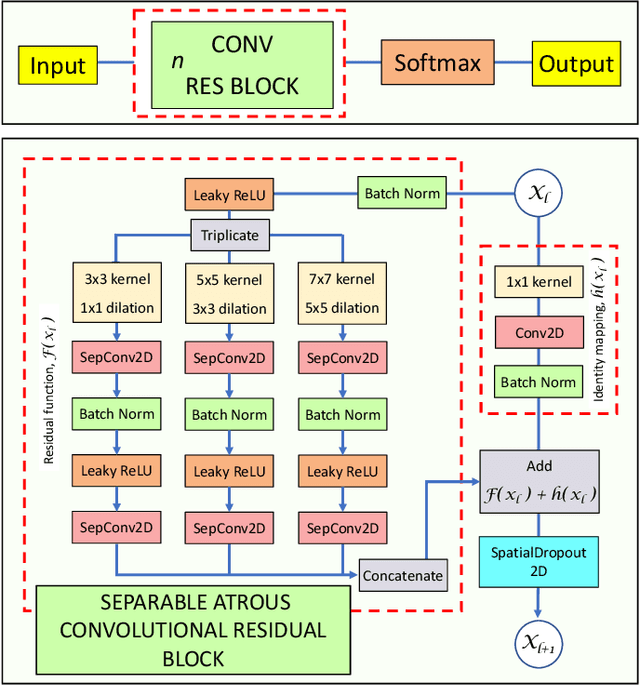
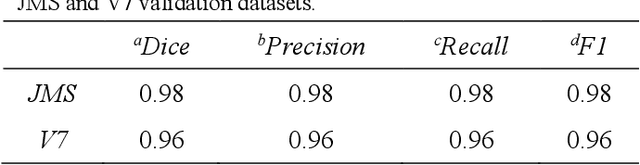
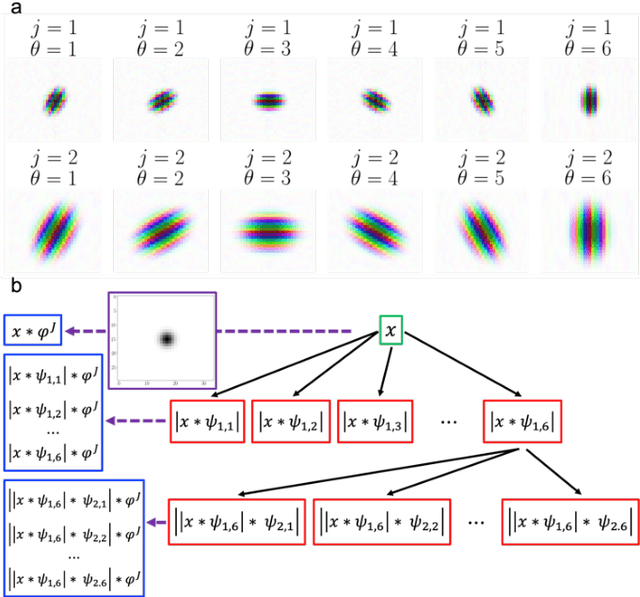
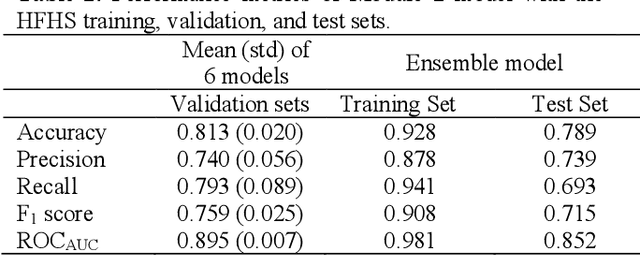
Abstract:CXR-Net is a two-module Artificial Intelligence pipeline for the quick detection of SARS-CoV-2 from chest X-rays (CXRs). Module 1 was trained on a public dataset of 6395 CXRs with radiologist annotated lung contours to generate masks of the lungs that overlap the heart and large vasa. Module 2 is a hybrid convnet in which the first convolutional layer with learned coefficients is replaced by a layer with fixed coefficients provided by the Wavelet Scattering Transform (WST). Module 2 takes as inputs the patients CXRs and corresponding lung masks calculated by Module 1, and produces as outputs a class assignment (Covid vs. non-Covid) and high resolution heat maps that identify the SARS associated lung regions. Module 2 was trained on a dataset of CXRs from non-Covid and RT-PCR confirmed Covid patients acquired at the Henry Ford Health System (HFHS) Hospital in Detroit. All non-Covid CXRs were from pre-Covid era (2018-2019), and included images from both normal lungs and lungs affected by non-Covid pathologies. Training and test sets consisted of 2265 CXRs (1417 Covid negative, 848 Covid positive), and 1532 CXRs (945 Covid negative, 587 Covid positive), respectively. Six distinct cross-validation models, each trained on 1887 images and validated against 378 images, were combined into an ensemble model that was used to classify the CXR images of the test set with resulting Accuracy = 0.789, Precision = 0.739, Recall = 0.693, F1 score = 0.715, ROC(AUC) = 0.852.
Lung Segmentation in Chest X-rays with Res-CR-Net
Nov 14, 2020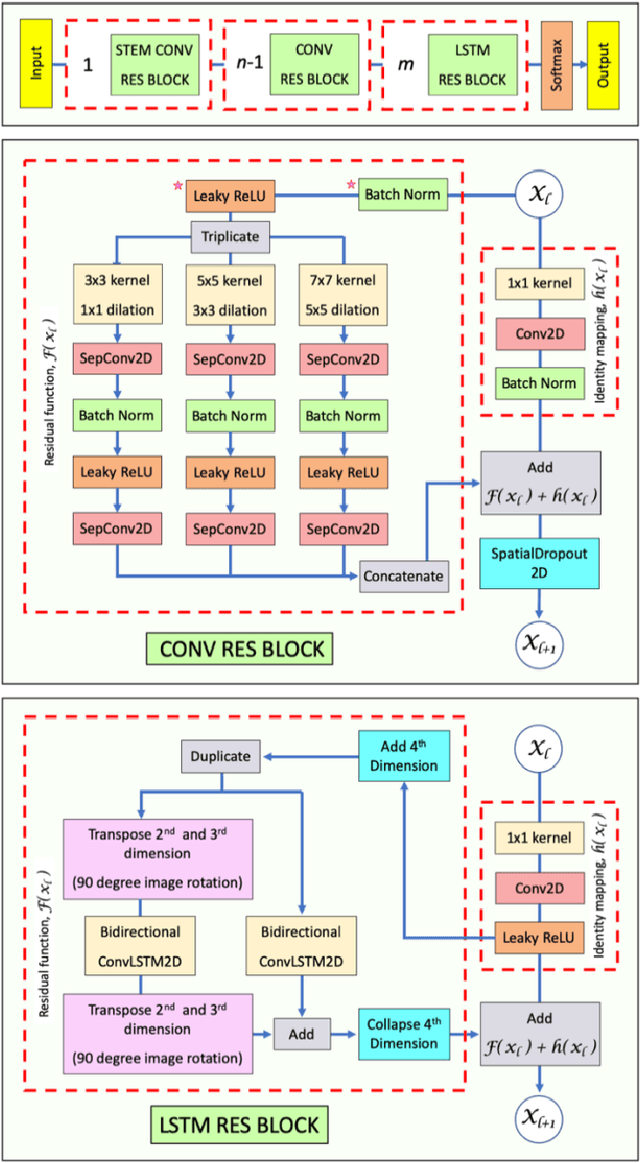
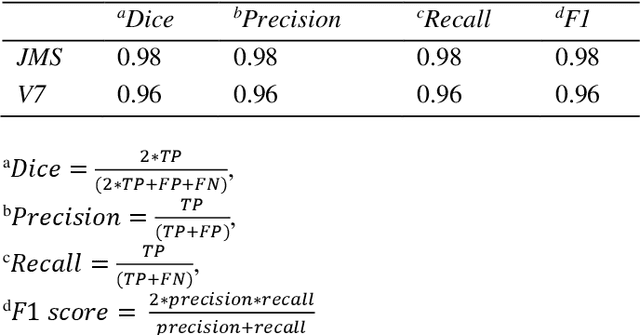
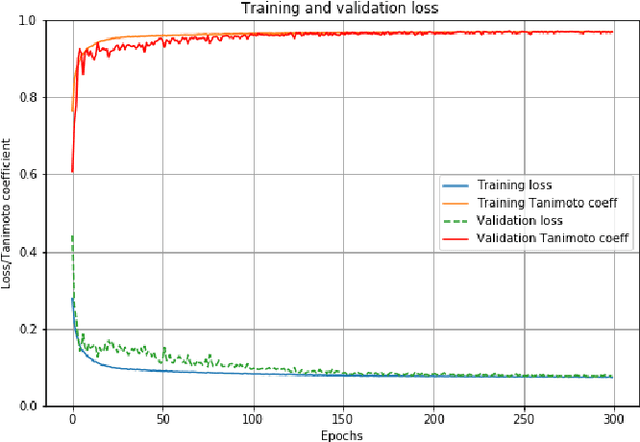
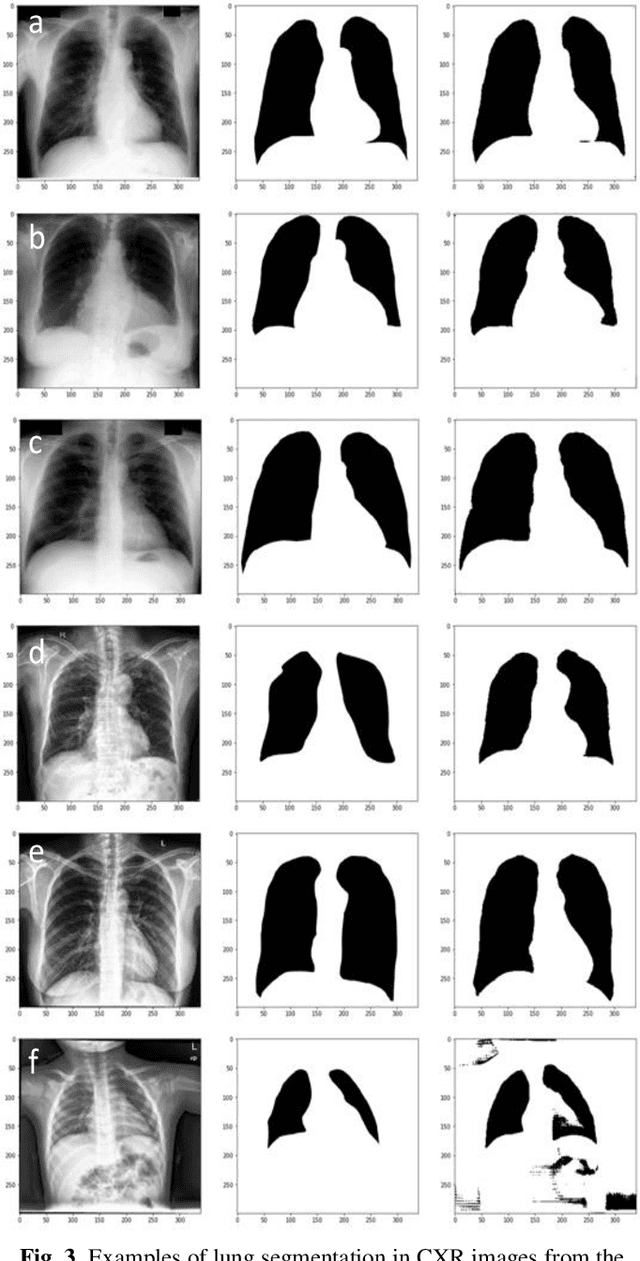
Abstract:Deep Neural Networks (DNN) are widely used to carry out segmentation tasks in biomedical images. Most DNNs developed for this purpose are based on some variation of the encoder-decoder U-Net architecture. Here we show that Res-CR-Net, a new type of fully convolutional neural network, which was originally developed for the semantic segmentation of microscopy images, and which does not adopt a U-Net architecture, is very effective at segmenting the lung fields in chest X-rays from either healthy patients or patients with a variety of lung pathologies.
Res-CR-Net, a residual network with a novel architecture optimized for the semantic segmentation of microscopy images
Apr 14, 2020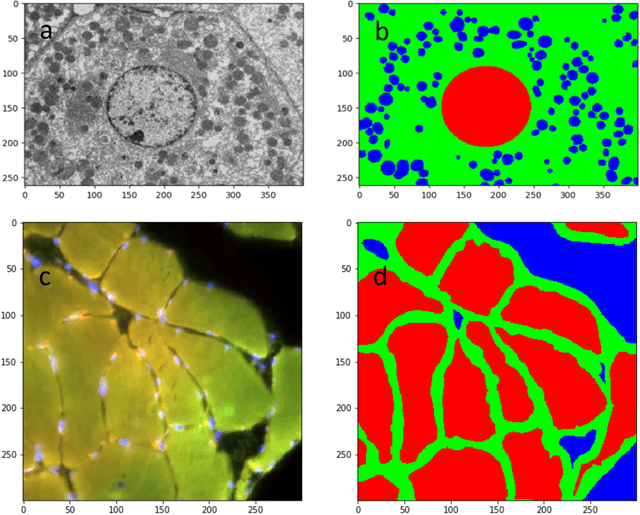
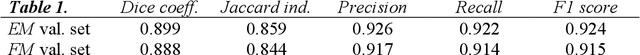
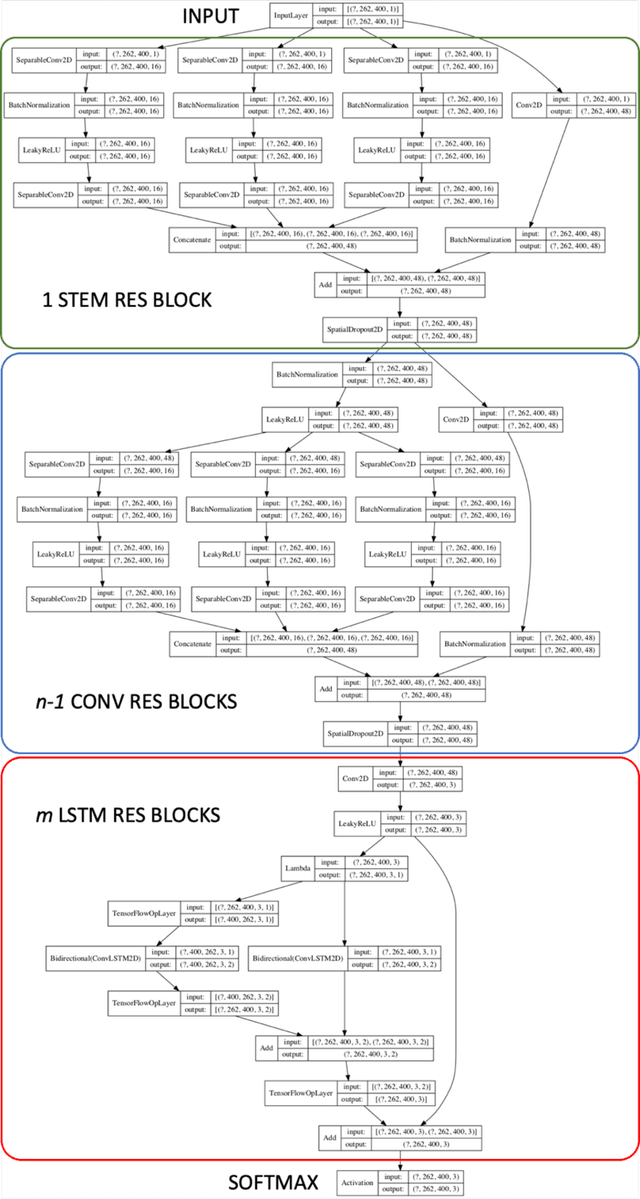
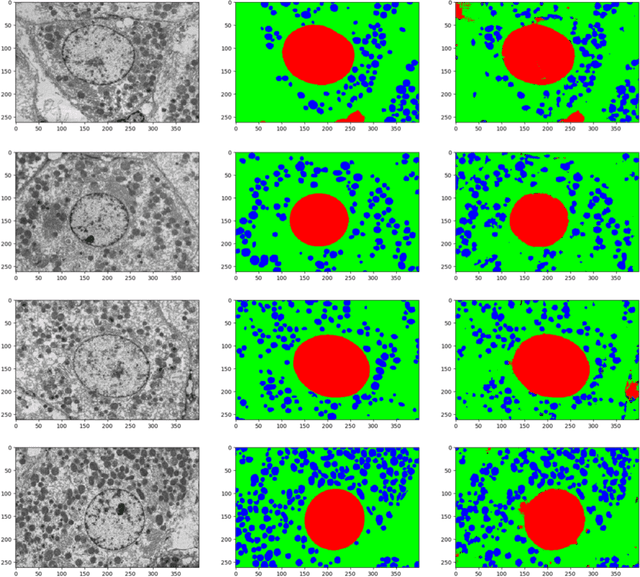
Abstract:Deep Neural Networks (DNN) have been widely used to carry out segmentation tasks in both electron and light microscopy. Most DNNs developed for this purpose are based on some variation of the encoder-decoder type U-Net architecture, in combination with residual blocks to increase ease of training and resilience to gradient degradation. Here we introduce Res-CR-Net, a type of DNN that features residual blocks with either a bundle of separable atrous convolutions with different dilation rates or a convolutional LSTM. The number of filters used in each residual block and the number of blocks are the only hyperparameters that need to be modified in order to optimize the network training for a variety of different microscopy images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge