Dmitry Minskiy
Subcellular Protein Localisation in the Human Protein Atlas using Ensembles of Diverse Deep Architectures
May 19, 2022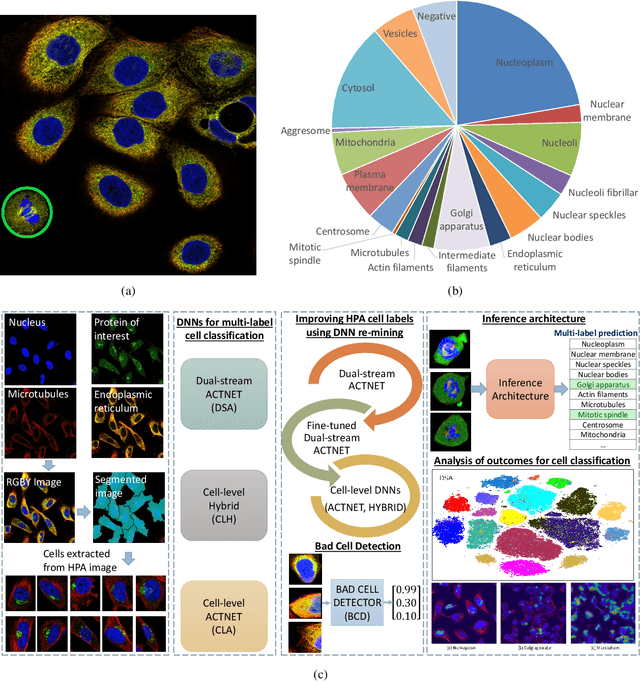
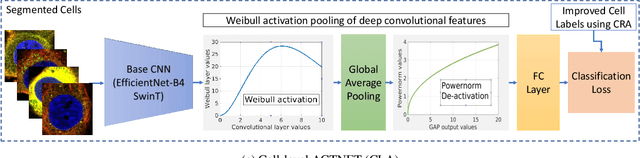
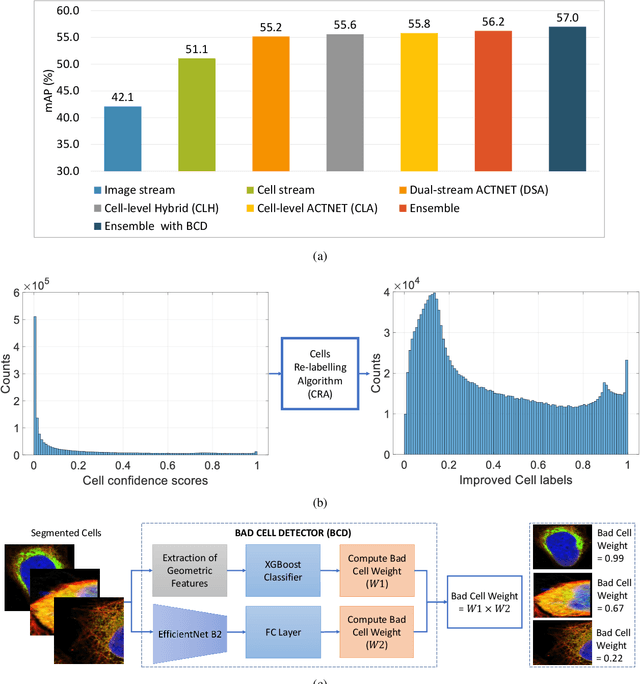
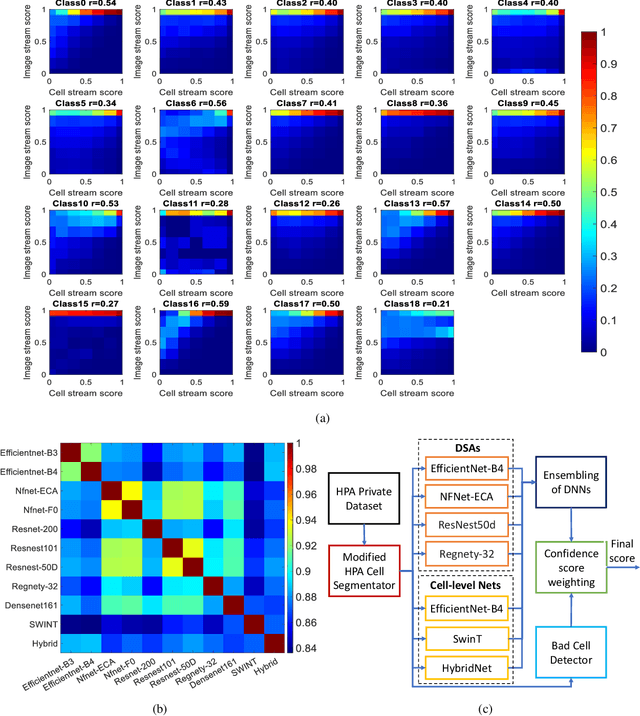
Abstract:Automated visual localisation of subcellular proteins can accelerate our understanding of cell function in health and disease. Despite recent advances in machine learning (ML), humans still attain superior accuracy by using diverse clues. We show how this gap can be narrowed by addressing three key aspects: (i) automated improvement of cell annotation quality, (ii) new Convolutional Neural Network (CNN) architectures supporting unbalanced and noisy data, and (iii) informed selection and fusion of multiple & diverse machine learning models. We introduce a new "AI-trains-AI" method for improving the quality of weak labels and propose novel CNN architectures exploiting wavelet filters and Weibull activations. We also explore key factors in the multi-CNN ensembling process by analysing correlations between image-level and cell-level predictions. Finally, in the context of the Human Protein Atlas, we demonstrate that our system achieves state-of-the-art performance in the multi-label single-cell classification of protein localisation patterns. It also significantly improves generalisation ability.
Efficient Hybrid Network: Inducting Scattering Features
Mar 29, 2022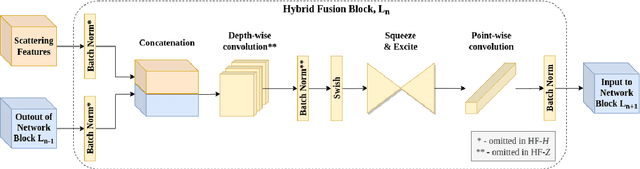
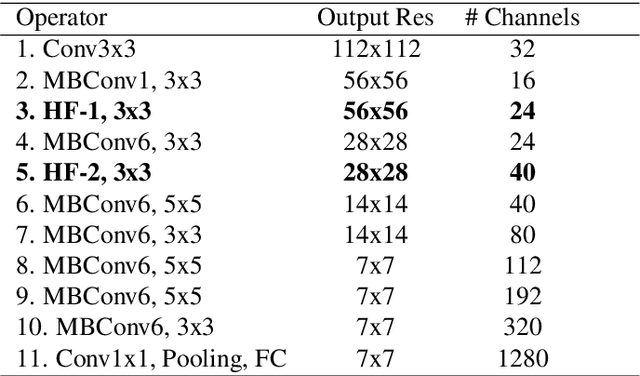
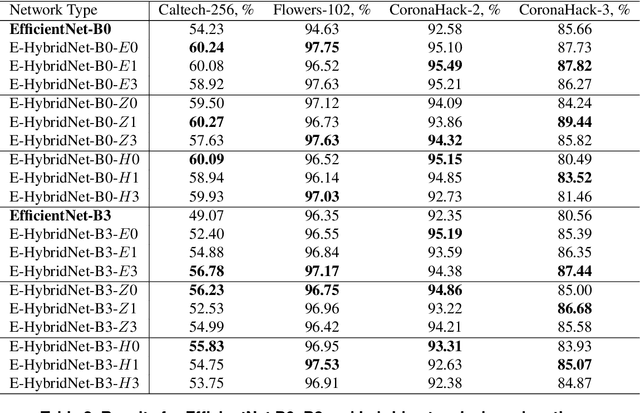
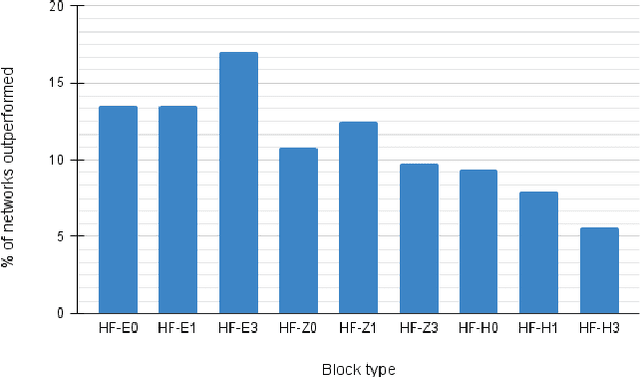
Abstract:Recent work showed that hybrid networks, which combine predefined and learnt filters within a single architecture, are more amenable to theoretical analysis and less prone to overfitting in data-limited scenarios. However, their performance has yet to prove competitive against the conventional counterparts when sufficient amounts of training data are available. In an attempt to address this core limitation of current hybrid networks, we introduce an Efficient Hybrid Network (E-HybridNet). We show that it is the first scattering based approach that consistently outperforms its conventional counterparts on a diverse range of datasets. It is achieved with a novel inductive architecture that embeds scattering features into the network flow using Hybrid Fusion Blocks. We also demonstrate that the proposed design inherits the key property of prior hybrid networks -- an effective generalisation in data-limited scenarios. Our approach successfully combines the best of the two worlds: flexibility and power of learnt features and stability and predictability of scattering representations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge