Dmitry Lachinov
Cephalometric Landmark Regression with Convolutional Neural Networks on 3D Computed Tomography Data
Jul 20, 2020
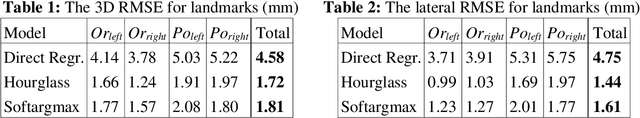
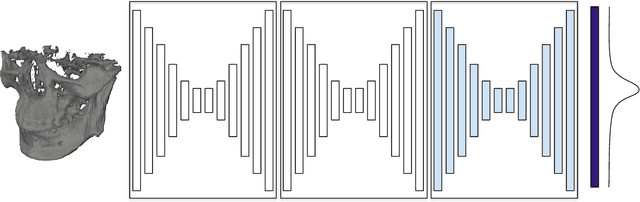
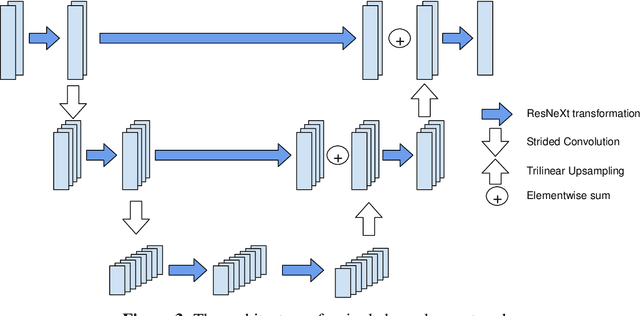
Abstract:In this paper, we address the problem of automatic three-dimensional cephalometric analysis. Cephalometric analysis performed on lateral radiographs doesn't fully exploit the structure of 3D objects due to projection onto the lateral plane. With the development of three-dimensional imaging techniques such as CT, several analysis methods have been proposed that extend to the 3D case. The analysis based on these methods is invariant to rotations and translations and can describe difficult skull deformation, where 2D cephalometry has no use. In this paper, we provide a wide overview of existing approaches for cephalometric landmark regression. Moreover, we perform a series of experiments with state of the art 3D convolutional neural network (CNN) based methods for keypoint regression: direct regression with CNN, heatmap regression and Softargmax regression. For the first time, we extensively evaluate the described methods and demonstrate their effectiveness in the estimation of Frankfort Horizontal and cephalometric points locations for patients with severe skull deformations. We demonstrate that Heatmap and Softargmax regression models provide sufficient regression error for medical applications (less than 4 mm). Moreover, the Softargmax model achieves 1.15o inclination error for the Frankfort horizontal. For the fair comparison with the prior art, we also report results projected on the lateral plane.
CHAOS Challenge -- Combined (CT-MR) Healthy Abdominal Organ Segmentation
Jan 17, 2020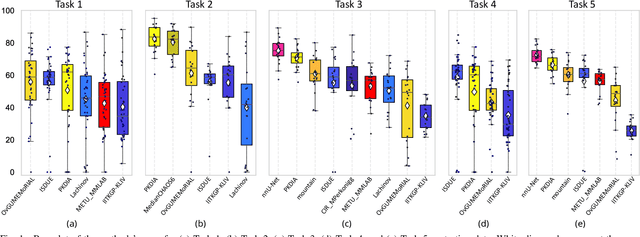
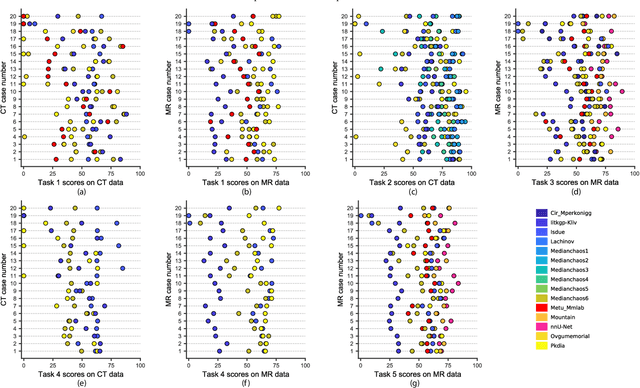
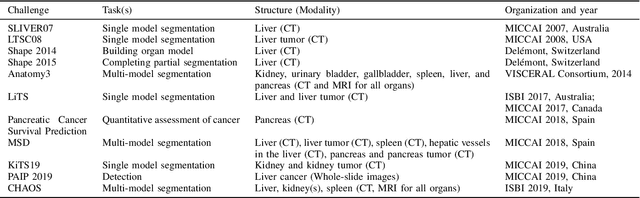
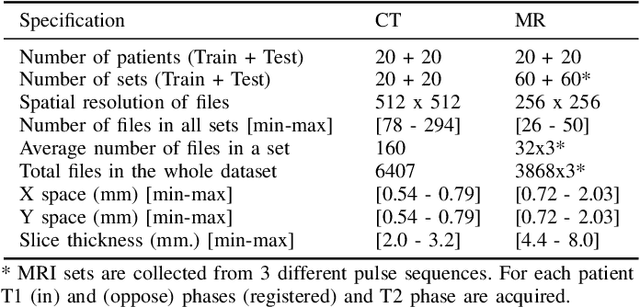
Abstract:Segmentation of abdominal organs has been a comprehensive, yet unresolved, research field for many years. In the last decade, intensive developments in deep learning (DL) have introduced new state-of-the-art segmentation systems. Despite outperforming the overall accuracy of existing systems, the effects of DL model properties and parameters on the performance is hard to interpret. This makes comparative analysis a necessary tool to achieve explainable studies and systems. Moreover, the performance of DL for emerging learning approaches such as cross-modality and multi-modal tasks have been rarely discussed. In order to expand the knowledge in these topics, CHAOS -- Combined (CT-MR) Healthy Abdominal Organ Segmentation challenge has been organized in the IEEE International Symposium on Biomedical Imaging (ISBI), 2019, in Venice, Italy. Despite a large number of the previous abdomen related challenges, the majority of which are focused on tumor/lesion detection and/or classification with a single modality, CHAOS provides both abdominal CT and MR data from healthy subjects. Five different and complementary tasks have been designed to analyze the capabilities of the current approaches from multiple perspectives. The results are investigated thoroughly, compared with manual annotations and interactive methods. The outcomes are reported in detail to reflect the latest advancements in the field. CHAOS challenge and data will be available online to provide a continuous benchmark resource for segmentation.
The Coherent Point Drift for Clustered Point Sets
Dec 14, 2018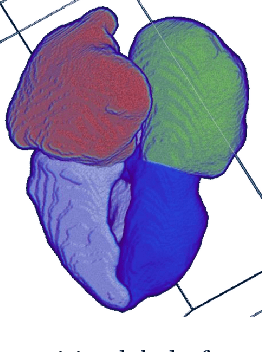

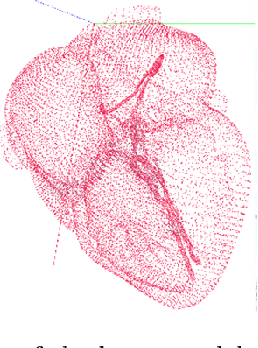
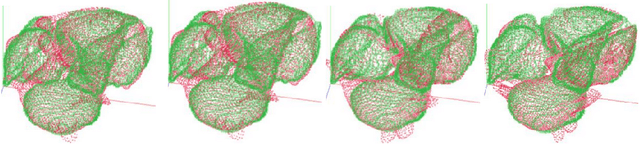
Abstract:The problem of non-rigid point set registration is a key problem for many computer vision tasks. In many cases the nature of the data or capabilities of the point detection algorithms can give us some prior information on point sets distribution. In non-rigid case this information is able to drastically improve registration results by limiting number of possible solutions. In this paper we explore use of prior information about point sets clustering, such information can be obtained with preliminary segmentation. We extend existing probabilistic framework for fitting two level Gaussian mixture model and derive closed form solution for maximization step of the EM algorithm. This enables us to improve method accuracy with almost no performance loss. We evaluate our approach and compare the Cluster Coherent Point Drift with other existing non-rigid point set registration methods and show it's advantages for digital medicine tasks, especially for heart template model personalization using patient's medical data.
Glioma Segmentation with Cascaded Unet
Oct 09, 2018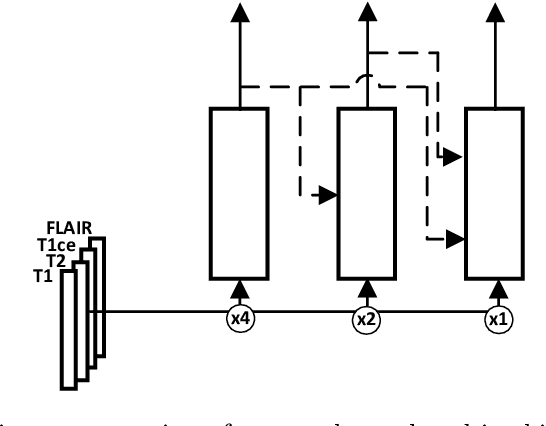
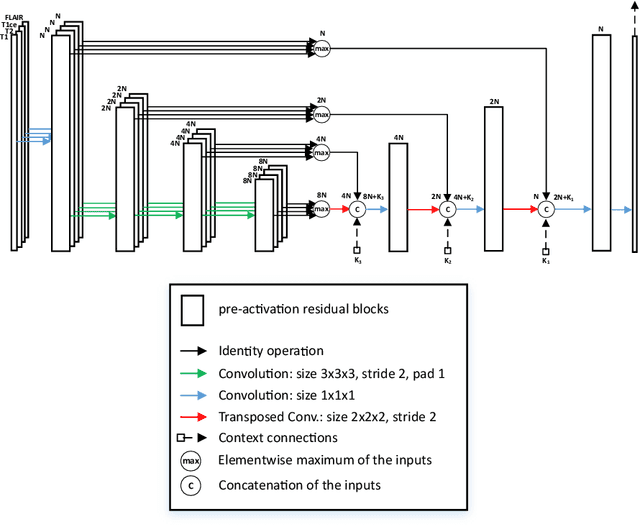
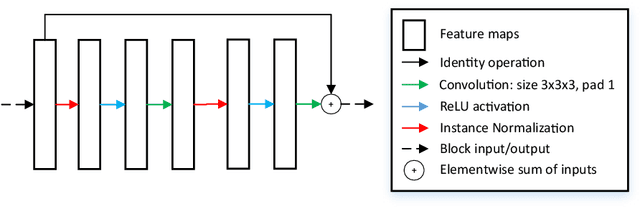
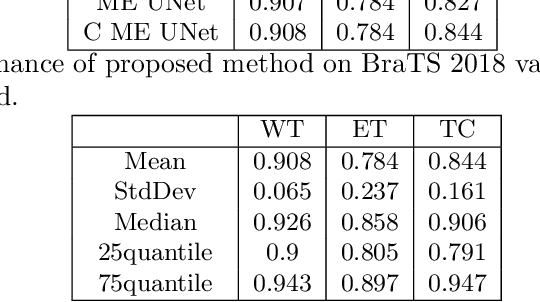
Abstract:MRI analysis takes central position in brain tumor diagnosis and treatment, thus it's precise evaluation is crucially important. However, it's 3D nature imposes several challenges, so the analysis is often performed on 2D projections that reduces the complexity, but increases bias. On the other hand, time consuming 3D evaluation, like, segmentation, is able to provide precise estimation of a number of valuable spatial characteristics, giving us understanding about the course of the disease.\newline Recent studies, focusing on the segmentation task, report superior performance of Deep Learning methods compared to classical computer vision algorithms. But still, it remains a challenging problem. In this paper we present deep cascaded approach for automatic brain tumor segmentation. Similar to recent methods for object detection, our implementation is based on neural networks; we propose modifications to the 3D UNet architecture and augmentation strategy to efficiently handle multimodal MRI input, besides this we introduce approach to enhance segmentation quality with context obtained from models of the same topology operating on downscaled data. We evaluate presented approach on BraTS 2018 dataset and discuss results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge