Vadim Turlapov
TUMLS: Trustful Fully Unsupervised Multi-Level Segmentation for Whole Slide Images of Histology
Apr 17, 2025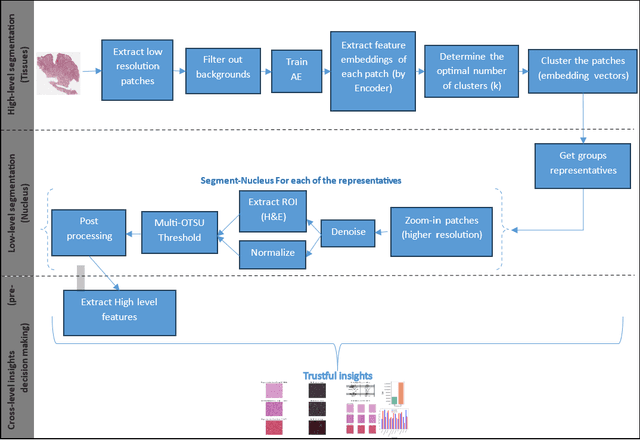
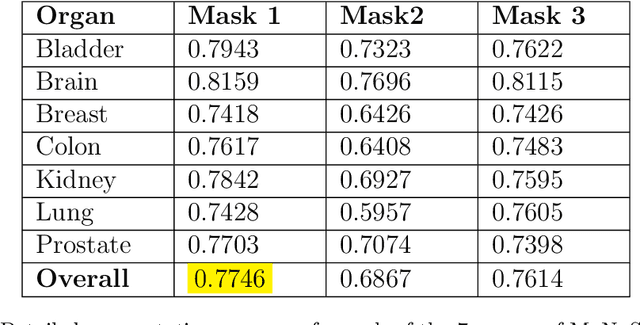
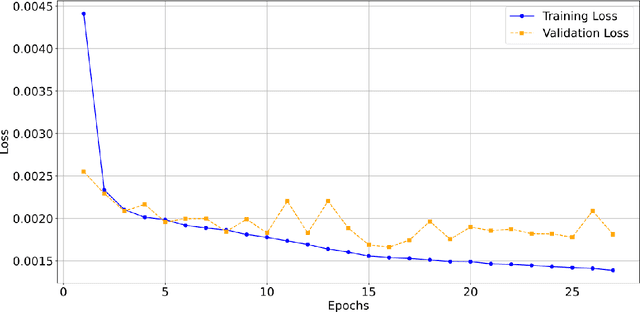
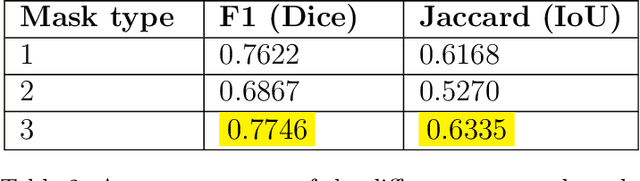
Abstract:Digital pathology, augmented by artificial intelligence (AI), holds significant promise for improving the workflow of pathologists. However, challenges such as the labor-intensive annotation of whole slide images (WSIs), high computational demands, and trust concerns arising from the absence of uncertainty estimation in predictions hinder the practical application of current AI methodologies in histopathology. To address these issues, we present a novel trustful fully unsupervised multi-level segmentation methodology (TUMLS) for WSIs. TUMLS adopts an autoencoder (AE) as a feature extractor to identify the different tissue types within low-resolution training data. It selects representative patches from each identified group based on an uncertainty measure and then does unsupervised nuclei segmentation in their respective higher-resolution space without using any ML algorithms. Crucially, this solution integrates seamlessly into clinicians workflows, transforming the examination of a whole WSI into a review of concise, interpretable cross-level insights. This integration significantly enhances and accelerates the workflow while ensuring transparency. We evaluated our approach using the UPENN-GBM dataset, where the AE achieved a mean squared error (MSE) of 0.0016. Additionally, nucleus segmentation is assessed on the MoNuSeg dataset, outperforming all unsupervised approaches with an F1 score of 77.46% and a Jaccard score of 63.35%. These results demonstrate the efficacy of TUMLS in advancing the field of digital pathology.
Cephalometric Landmark Regression with Convolutional Neural Networks on 3D Computed Tomography Data
Jul 20, 2020
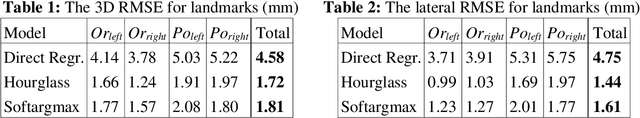
Abstract:In this paper, we address the problem of automatic three-dimensional cephalometric analysis. Cephalometric analysis performed on lateral radiographs doesn't fully exploit the structure of 3D objects due to projection onto the lateral plane. With the development of three-dimensional imaging techniques such as CT, several analysis methods have been proposed that extend to the 3D case. The analysis based on these methods is invariant to rotations and translations and can describe difficult skull deformation, where 2D cephalometry has no use. In this paper, we provide a wide overview of existing approaches for cephalometric landmark regression. Moreover, we perform a series of experiments with state of the art 3D convolutional neural network (CNN) based methods for keypoint regression: direct regression with CNN, heatmap regression and Softargmax regression. For the first time, we extensively evaluate the described methods and demonstrate their effectiveness in the estimation of Frankfort Horizontal and cephalometric points locations for patients with severe skull deformations. We demonstrate that Heatmap and Softargmax regression models provide sufficient regression error for medical applications (less than 4 mm). Moreover, the Softargmax model achieves 1.15o inclination error for the Frankfort horizontal. For the fair comparison with the prior art, we also report results projected on the lateral plane.
Knowledge Distillation for Brain Tumor Segmentation
Feb 10, 2020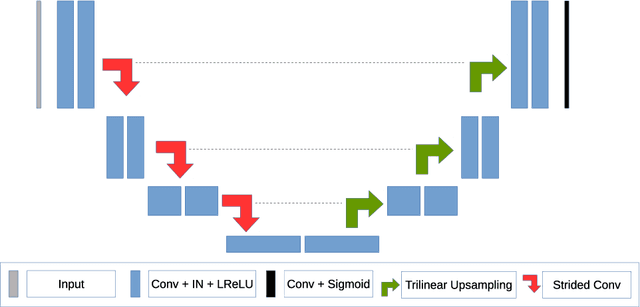
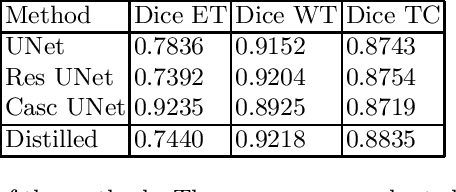
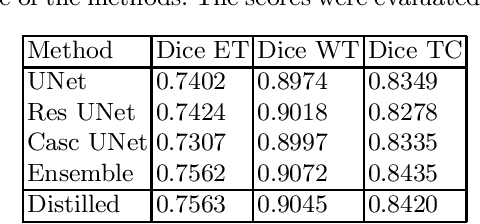
Abstract:The segmentation of brain tumors in multimodal MRIs is one of the most challenging tasks in medical image analysis. The recent state of the art algorithms solving this task is based on machine learning approaches and deep learning in particular. The amount of data used for training such models and its variability is a keystone for building an algorithm with high representation power. In this paper, we study the relationship between the performance of the model and the amount of data employed during the training process. On the example of brain tumor segmentation challenge, we compare the model trained with labeled data provided by challenge organizers, and the same model trained in omni-supervised manner using additional unlabeled data annotated with the ensemble of heterogeneous models. As a result, a single model trained with additional data achieves performance close to the ensemble of multiple models and outperforms individual methods.
The Coherent Point Drift for Clustered Point Sets
Dec 14, 2018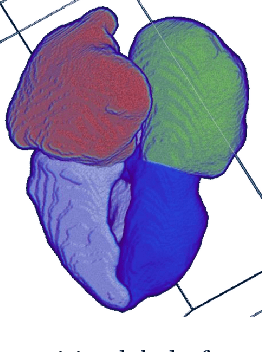

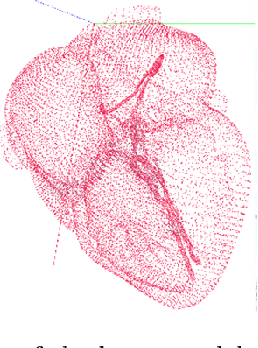
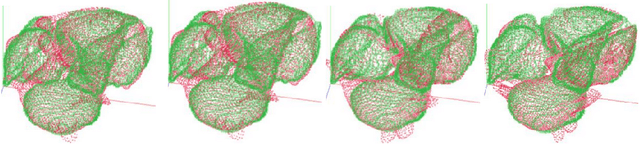
Abstract:The problem of non-rigid point set registration is a key problem for many computer vision tasks. In many cases the nature of the data or capabilities of the point detection algorithms can give us some prior information on point sets distribution. In non-rigid case this information is able to drastically improve registration results by limiting number of possible solutions. In this paper we explore use of prior information about point sets clustering, such information can be obtained with preliminary segmentation. We extend existing probabilistic framework for fitting two level Gaussian mixture model and derive closed form solution for maximization step of the EM algorithm. This enables us to improve method accuracy with almost no performance loss. We evaluate our approach and compare the Cluster Coherent Point Drift with other existing non-rigid point set registration methods and show it's advantages for digital medicine tasks, especially for heart template model personalization using patient's medical data.
Glioma Segmentation with Cascaded Unet
Oct 09, 2018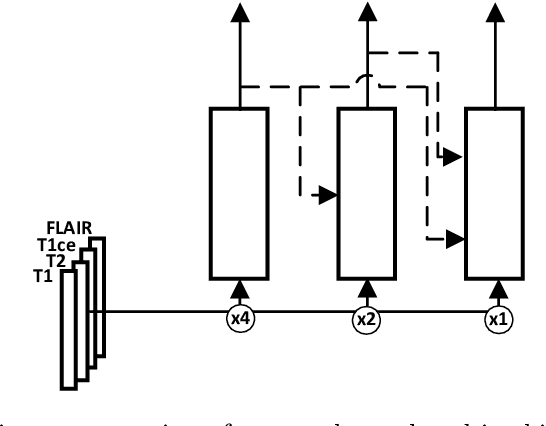
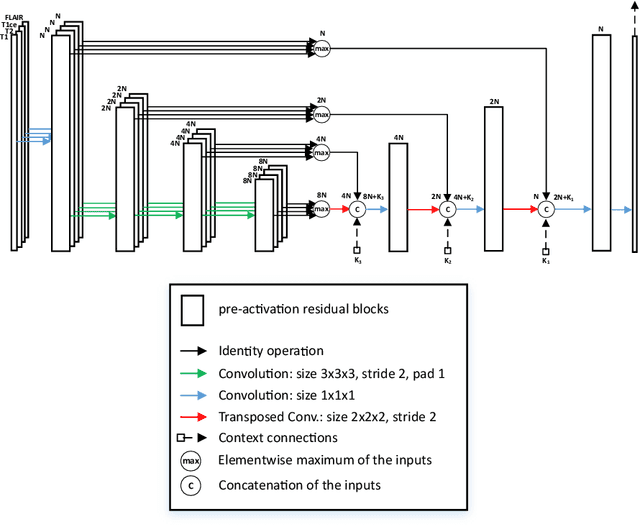
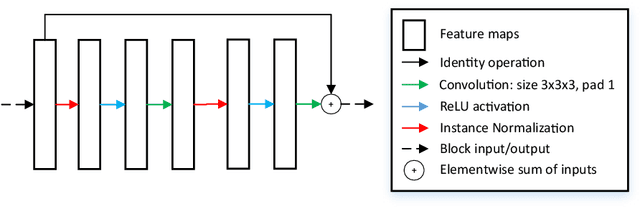
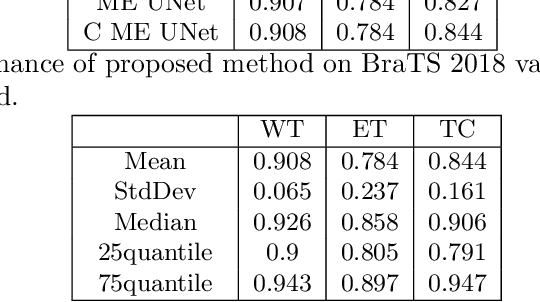
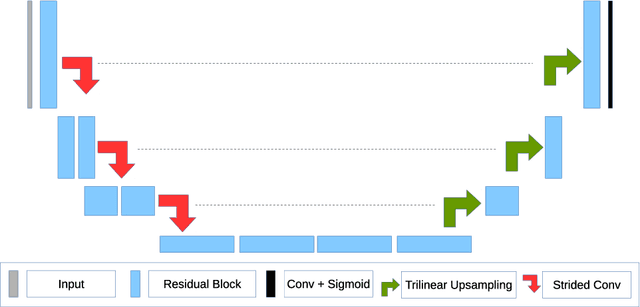
Abstract:MRI analysis takes central position in brain tumor diagnosis and treatment, thus it's precise evaluation is crucially important. However, it's 3D nature imposes several challenges, so the analysis is often performed on 2D projections that reduces the complexity, but increases bias. On the other hand, time consuming 3D evaluation, like, segmentation, is able to provide precise estimation of a number of valuable spatial characteristics, giving us understanding about the course of the disease.\newline Recent studies, focusing on the segmentation task, report superior performance of Deep Learning methods compared to classical computer vision algorithms. But still, it remains a challenging problem. In this paper we present deep cascaded approach for automatic brain tumor segmentation. Similar to recent methods for object detection, our implementation is based on neural networks; we propose modifications to the 3D UNet architecture and augmentation strategy to efficiently handle multimodal MRI input, besides this we introduce approach to enhance segmentation quality with context obtained from models of the same topology operating on downscaled data. We evaluate presented approach on BraTS 2018 dataset and discuss results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge