Dimitris Margaritis
Efficient Markov Network Structure Discovery Using Independence Tests
Jan 15, 2014
Abstract:We present two algorithms for learning the structure of a Markov network from data: GSMN* and GSIMN. Both algorithms use statistical independence tests to infer the structure by successively constraining the set of structures consistent with the results of these tests. Until very recently, algorithms for structure learning were based on maximum likelihood estimation, which has been proved to be NP-hard for Markov networks due to the difficulty of estimating the parameters of the network, needed for the computation of the data likelihood. The independence-based approach does not require the computation of the likelihood, and thus both GSMN* and GSIMN can compute the structure efficiently (as shown in our experiments). GSMN* is an adaptation of the Grow-Shrink algorithm of Margaritis and Thrun for learning the structure of Bayesian networks. GSIMN extends GSMN* by additionally exploiting Pearls well-known properties of the conditional independence relation to infer novel independences from known ones, thus avoiding the performance of statistical tests to estimate them. To accomplish this efficiently GSIMN uses the Triangle theorem, also introduced in this work, which is a simplified version of the set of Markov axioms. Experimental comparisons on artificial and real-world data sets show GSIMN can yield significant savings with respect to GSMN*, while generating a Markov network with comparable or in some cases improved quality. We also compare GSIMN to a forward-chaining implementation, called GSIMN-FCH, that produces all possible conditional independences resulting from repeatedly applying Pearls theorems on the known conditional independence tests. The results of this comparison show that GSIMN, by the sole use of the Triangle theorem, is nearly optimal in terms of the set of independences tests that it infers.
A Bayesian Multiresolution Independence Test for Continuous Variables
Jan 10, 2013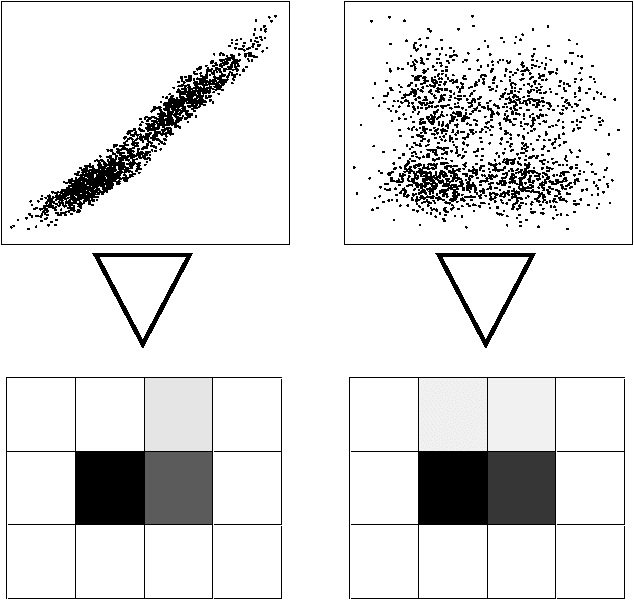

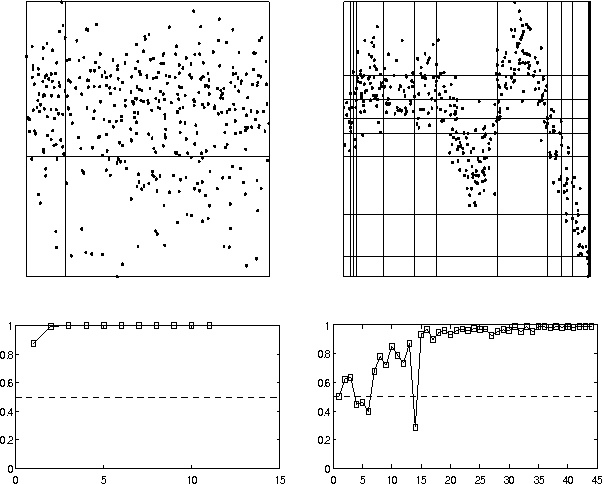
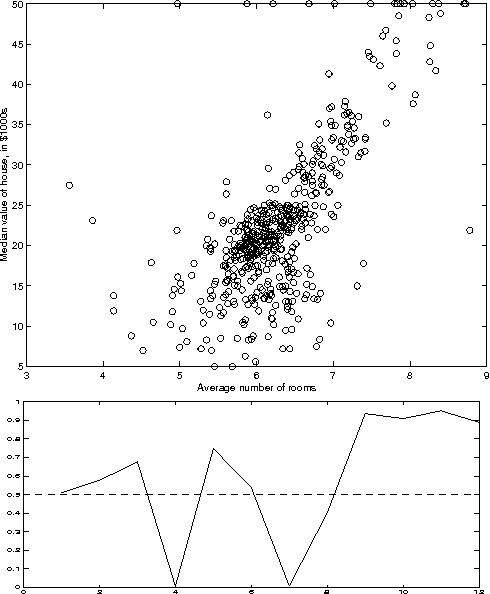
Abstract:In this paper we present a method ofcomputing the posterior probability ofconditional independence of two or morecontinuous variables from data,examined at several resolutions. Ourapproach is motivated by theobservation that the appearance ofcontinuous data varies widely atvarious resolutions, producing verydifferent independence estimatesbetween the variablesinvolved. Therefore, it is difficultto ascertain independence withoutexamining data at several carefullyselected resolutions. In our paper, weaccomplish this using the exactcomputation of the posteriorprobability of independence, calculatedanalytically given a resolution. Ateach examined resolution, we assume amultinomial distribution with Dirichletpriors for the discretized tableparameters, and compute the posteriorusing Bayesian integration. Acrossresolutions, we use a search procedureto approximate the Bayesian integral ofprobability over an exponential numberof possible histograms. Our methodgeneralizes to an arbitrary numbervariables in a straightforward manner.The test is suitable for Bayesiannetwork learning algorithms that useindependence tests to infer the networkstructure, in domains that contain anymix of continuous, ordinal andcategorical variables.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge