Diego Molla-Aliod
LLM Ensemble for RAG: Role of Context Length in Zero-Shot Question Answering for BioASQ Challenge
Sep 10, 2025



Abstract:Biomedical question answering (QA) poses significant challenges due to the need for precise interpretation of specialized knowledge drawn from a vast, complex, and rapidly evolving corpus. In this work, we explore how large language models (LLMs) can be used for information retrieval (IR), and an ensemble of zero-shot models can accomplish state-of-the-art performance on a domain-specific Yes/No QA task. Evaluating our approach on the BioASQ challenge tasks, we show that ensembles can outperform individual LLMs and in some cases rival or surpass domain-tuned systems - all while preserving generalizability and avoiding the need for costly fine-tuning or labeled data. Our method aggregates outputs from multiple LLM variants, including models from Anthropic and Google, to synthesize more accurate and robust answers. Moreover, our investigation highlights a relationship between context length and performance: while expanded contexts are meant to provide valuable evidence, they simultaneously risk information dilution and model disorientation. These findings emphasize IR as a critical foundation in Retrieval-Augmented Generation (RAG) approaches for biomedical QA systems. Precise, focused retrieval remains essential for ensuring LLMs operate within relevant information boundaries when generating answers from retrieved documents. Our results establish that ensemble-based zero-shot approaches, when paired with effective RAG pipelines, constitute a practical and scalable alternative to domain-tuned systems for biomedical question answering.
Synthetic Dialogue Dataset Generation using LLM Agents
Jan 30, 2024

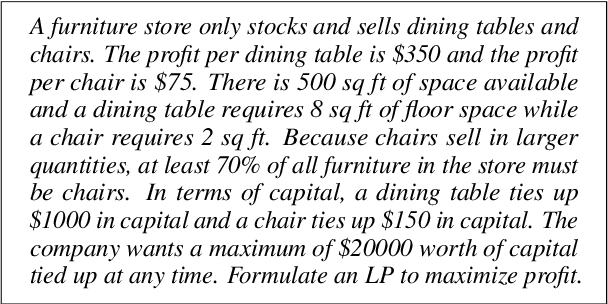
Abstract:Linear programming (LP) problems are pervasive in real-life applications. However, despite their apparent simplicity, an untrained user may find it difficult to determine the linear model of their specific problem. We envisage the creation of a goal-oriented conversational agent that will engage in conversation with the user to elicit all information required so that a subsequent agent can generate the linear model. In this paper, we present an approach for the generation of sample dialogues that can be used to develop and train such a conversational agent. Using prompt engineering, we develop two agents that "talk" to each other, one acting as the conversational agent, and the other acting as the user. Using a set of text descriptions of linear problems from NL4Opt available to the user only, the agent and the user engage in conversation until the agent has retrieved all key information from the original problem description. We also propose an extrinsic evaluation of the dialogues by assessing how well the summaries generated by the dialogues match the original problem descriptions. We conduct human and automatic evaluations, including an evaluation approach that uses GPT-4 to mimic the human evaluation metrics. The evaluation results show an overall good quality of the dialogues, though research is still needed to improve the quality of the GPT-4 evaluation metrics. The resulting dialogues, including the human annotations of a subset, are available to the research community. The conversational agent used for the generation of the dialogues can be used as a baseline.
On Extending Neural Networks with Loss Ensembles for Text Classification
Nov 14, 2017
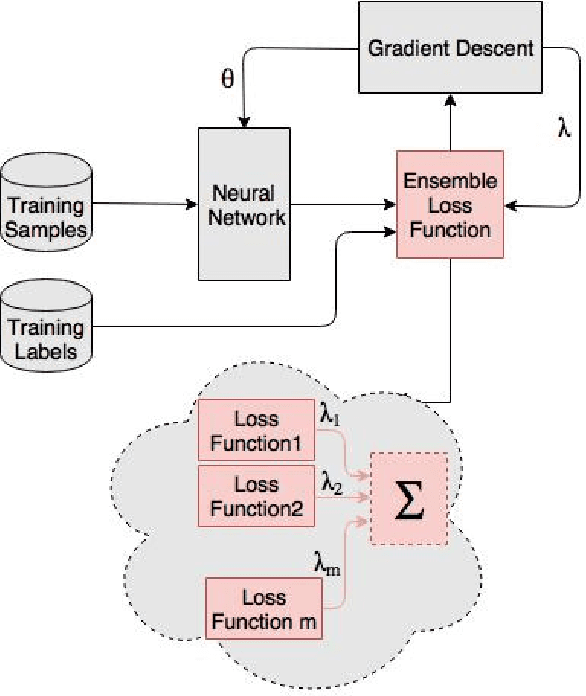


Abstract:Ensemble techniques are powerful approaches that combine several weak learners to build a stronger one. As a meta learning framework, ensemble techniques can easily be applied to many machine learning techniques. In this paper we propose a neural network extended with an ensemble loss function for text classification. The weight of each weak loss function is tuned within the training phase through the gradient propagation optimization method of the neural network. The approach is evaluated on several text classification datasets. We also evaluate its performance in various environments with several degrees of label noise. Experimental results indicate an improvement of the results and strong resilience against label noise in comparison with other methods.
Macquarie University at BioASQ 5b -- Query-based Summarisation Techniques for Selecting the Ideal Answers
Aug 11, 2017



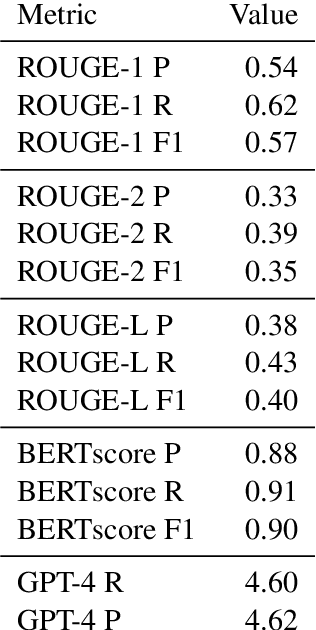
Abstract:Macquarie University's contribution to the BioASQ challenge (Task 5b Phase B) focused on the use of query-based extractive summarisation techniques for the generation of the ideal answers. Four runs were submitted, with approaches ranging from a trivial system that selected the first $n$ snippets, to the use of deep learning approaches under a regression framework. Our experiments and the ROUGE results of the five test batches of BioASQ indicate surprisingly good results for the trivial approach. Overall, most of our runs on the first three test batches achieved the best ROUGE-SU4 results in the challenge.
* As published in BioNLP2017. 9 pages, 5 figures, 4 tables
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge