Diego Mollá
Macquarie University, Sydney, Australia
Overview of the 2024 ALTA Shared Task: Detect Automatic AI-Generated Sentences for Human-AI Hybrid Articles
Dec 19, 2024


Abstract:The ALTA shared tasks have been running annually since 2010. In 2024, the purpose of the task is to detect machine-generated text in a hybrid setting where the text may contain portions of human text and portions machine-generated. In this paper, we present the task, the evaluation criteria, and the results of the systems participating in the shared task.
* 6 pages, 3 tables, published in ALTA 2024
Large Language Models and Prompt Engineering for Biomedical Query Focused Multi-Document Summarisation
Nov 09, 2023Abstract:This paper reports on the use of prompt engineering and GPT-3.5 for biomedical query-focused multi-document summarisation. Using GPT-3.5 and appropriate prompts, our system achieves top ROUGE-F1 results in the task of obtaining short-paragraph-sized answers to biomedical questions in the 2023 BioASQ Challenge (BioASQ 11b). This paper confirms what has been observed in other domains: 1) Prompts that incorporated few-shot samples generally improved on their counterpart zero-shot variants; 2) The largest improvement was achieved by retrieval augmented generation. The fact that these prompts allow our top runs to rank within the top two runs of BioASQ 11b demonstrate the power of using adequate prompts for Large Language Models in general, and GPT-3.5 in particular, for query-focused summarisation.
Query-focused Extractive Summarisation for Biomedical and COVID-19 Complex Question Answering
Sep 05, 2022



Abstract:This paper presents Macquarie University's participation to the two most recent BioASQ Synergy Tasks (as per June 2022), and to the BioASQ10 Task~B (BioASQ10b), Phase~B. In these tasks, participating systems are expected to generate complex answers to biomedical questions, where the answers may contain more than one sentence. We apply query-focused extractive summarisation techniques. In particular, we follow a sentence classification-based approach that scores each candidate sentence associated to a question, and the $n$ highest-scoring sentences are returned as the answer. The Synergy Task corresponds to an end-to-end system that requires document selection, snippet selection, and finding the final answer, but it has very limited training data. For the Synergy task, we selected the candidate sentences following two phases: document retrieval and snippet retrieval, and the final answer was found by using a DistilBERT/ALBERT classifier that had been trained on the training data of BioASQ9b. Document retrieval was achieved as a standard search over the CORD-19 data using the search API provided by the BioASQ organisers, and snippet retrieval was achieved by re-ranking the sentences of the top retrieved documents, using the cosine similarity of the question and candidate sentence. We observed that vectors represented via sBERT have an edge over tf.idf. BioASQ10b Phase B focuses on finding the specific answers to biomedical questions. For this task, we followed a data-centric approach. We hypothesised that the training data of the first BioASQ years might be biased and we experimented with different subsets of the training data. We observed an improvement of results when the system was trained on the second half of the BioASQ10b training data.
Transformer-based Language Models for Factoid Question Answering at BioASQ9b
Sep 15, 2021



Abstract:In this work, we describe our experiments and participating systems in the BioASQ Task 9b Phase B challenge of biomedical question answering. We have focused on finding the ideal answers and investigated multi-task fine-tuning and gradual unfreezing techniques on transformer-based language models. For factoid questions, our ALBERT-based systems ranked first in test batch 1 and fourth in test batch 2. Our DistilBERT systems outperformed the ALBERT variants in test batches 4 and 5 despite having 81% fewer parameters than ALBERT. However, we observed that gradual unfreezing had no significant impact on the model's accuracy compared to standard fine-tuning.
Query-Focused Extractive Summarisation for Finding Ideal Answers to Biomedical and COVID-19 Questions
Aug 31, 2021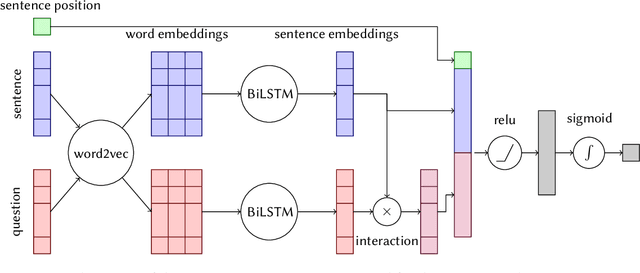
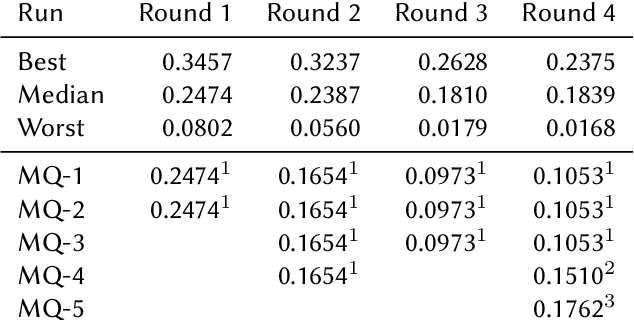
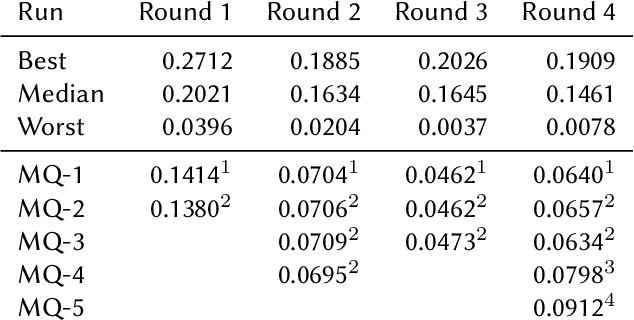
Abstract:This paper presents Macquarie University's participation to the BioASQ Synergy Task, and BioASQ9b Phase B. In each of these tasks, our participation focused on the use of query-focused extractive summarisation to obtain the ideal answers to medical questions. The Synergy Task is an end-to-end question answering task on COVID-19 where systems are required to return relevant documents, snippets, and answers to a given question. Given the absence of training data, we used a query-focused summarisation system that was trained with the BioASQ8b training data set and we experimented with methods to retrieve the documents and snippets. Considering the poor quality of the documents and snippets retrieved by our system, we observed reasonably good quality in the answers returned. For phase B of the BioASQ9b task, the relevant documents and snippets were already included in the test data. Our system split the snippets into candidate sentences and used BERT variants under a sentence classification setup. The system used the question and candidate sentence as input and was trained to predict the likelihood of the candidate sentence being part of the ideal answer. The runs obtained either the best or second best ROUGE-F1 results of all participants to all batches of BioASQ9b. This shows that using BERT in a classification setup is a very strong baseline for the identification of ideal answers.
Macquarie University at BioASQ 6b: Deep learning and deep reinforcement learning for query-based multi-document summarisation
Sep 14, 2018



Abstract:This paper describes Macquarie University's contribution to the BioASQ Challenge (BioASQ 6b, Phase B). We focused on the extraction of the ideal answers, and the task was approached as an instance of query-based multi-document summarisation. In particular, this paper focuses on the experiments related to the deep learning and reinforcement learning approaches used in the submitted runs. The best run used a deep learning model under a regression-based framework. The deep learning architecture used features derived from the output of LSTM chains on word embeddings, plus features based on similarity with the query, and sentence position. The reinforcement learning approach was a proof-of-concept prototype that trained a global policy using REINFORCE. The global policy was implemented as a neural network that used $tf.idf$ features encoding the candidate sentence, question, and context.
Supervised Machine Learning for Extractive Query Based Summarisation of Biomedical Data
Sep 14, 2018



Abstract:The automation of text summarisation of biomedical publications is a pressing need due to the plethora of information available on-line. This paper explores the impact of several supervised machine learning approaches for extracting multi-document summaries for given queries. In particular, we compare classification and regression approaches for query-based extractive summarisation using data provided by the BioASQ Challenge. We tackled the problem of annotating sentences for training classification systems and show that a simple annotation approach outperforms regression-based summarisation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge