Query-Focused Extractive Summarisation for Finding Ideal Answers to Biomedical and COVID-19 Questions
Paper and Code
Aug 31, 2021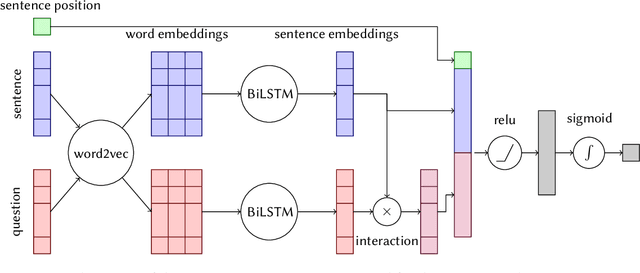
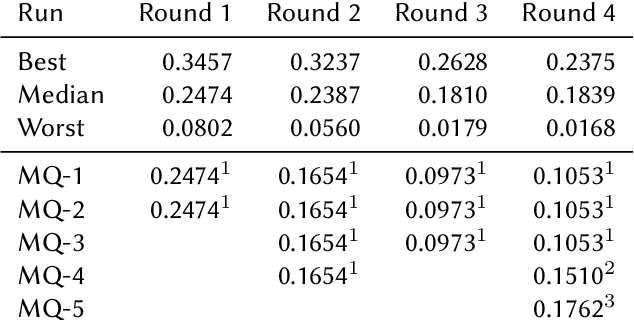
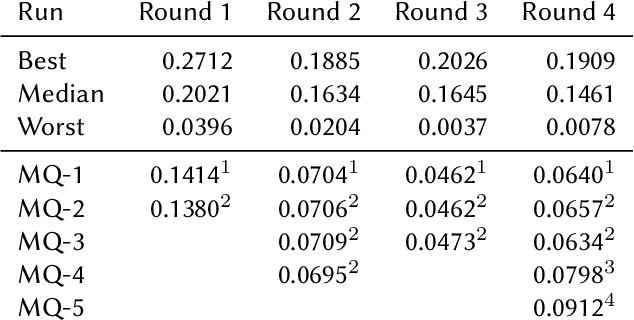
This paper presents Macquarie University's participation to the BioASQ Synergy Task, and BioASQ9b Phase B. In each of these tasks, our participation focused on the use of query-focused extractive summarisation to obtain the ideal answers to medical questions. The Synergy Task is an end-to-end question answering task on COVID-19 where systems are required to return relevant documents, snippets, and answers to a given question. Given the absence of training data, we used a query-focused summarisation system that was trained with the BioASQ8b training data set and we experimented with methods to retrieve the documents and snippets. Considering the poor quality of the documents and snippets retrieved by our system, we observed reasonably good quality in the answers returned. For phase B of the BioASQ9b task, the relevant documents and snippets were already included in the test data. Our system split the snippets into candidate sentences and used BERT variants under a sentence classification setup. The system used the question and candidate sentence as input and was trained to predict the likelihood of the candidate sentence being part of the ideal answer. The runs obtained either the best or second best ROUGE-F1 results of all participants to all batches of BioASQ9b. This shows that using BERT in a classification setup is a very strong baseline for the identification of ideal answers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge