Dennis Hedderich
Stochastic Cortical Self-Reconstruction
Mar 11, 2024Abstract:Magnetic resonance imaging (MRI) is critical for diagnosing neurodegenerative diseases, yet accurately assessing mild cortical atrophy remains a challenge due to its subtlety. Automated cortex reconstruction, paired with healthy reference ranges, aids in pinpointing pathological atrophy, yet their generalization is limited by biases from image acquisition and processing. We introduce the concept of stochastic cortical self-reconstruction (SCSR) that creates a subject-specific healthy reference by taking MRI-derived thicknesses as input and, therefore, implicitly accounting for potential confounders. SCSR randomly corrupts parts of the cortex and self-reconstructs them from the remaining information. Trained exclusively on healthy individuals, repeated self-reconstruction generates a stochastic reference cortex for assessing deviations from the norm. We present three implementations of this concept: XGBoost applied on parcels, and two autoencoders on vertex level -- one based on a multilayer perceptron and the other using a spherical U-Net. These models were trained on healthy subjects from the UK Biobank and subsequently evaluated across four public Alzheimer's datasets. Finally, we deploy the model on clinical in-house data, where deviation maps' high spatial resolution aids in discriminating between four types of dementia.
Complex Grey Matter Structure Segmentation in Brains via Deep Learning: Example of the Claustrum
Aug 08, 2020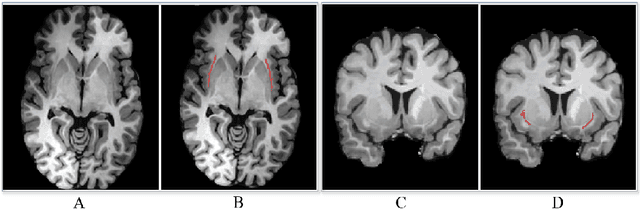
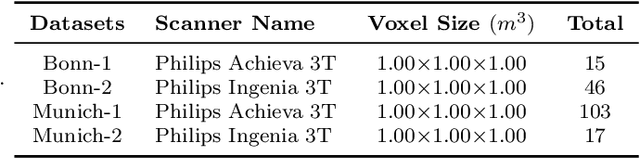
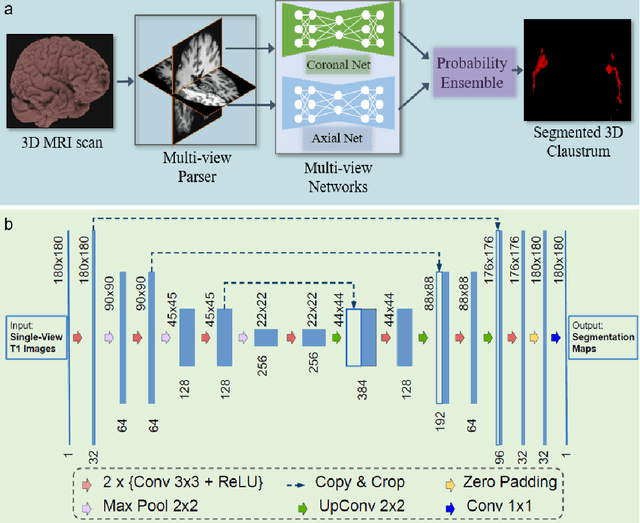

Abstract:Segmentationand parcellation of the brain has been widely performed on brain MRI using atlas-based methods. However, segmentation of the claustrum, a thin and sheet-like structure between insular cortex and putamen has not been amenable to automatized segmentation, thus limiting its investigation in larger imaging cohorts. Recently, deep-learning based approaches have been introduced for automated segmentation of brain structures, yielding great potential to overcome preexisting limitations. In the following, we present a multi-view deep-learning based approach to segment the claustrum in T1-weighted MRI scans. We trained and evaluated the proposed method on 181 manual bilateral claustrum annotations by an expert neuroradiologist serving as reference standard. Cross-validation experiments yielded median volumetric similarity, robust Hausdor? distance and Dice score of 93.3%, 1.41mm and 71.8% respectively which represents equal or superior segmentation performance compared to human intra-rater reliability. Leave-one-scanner-out evaluation showed good transfer-ability of the algorithm to images from unseen scanners, however at slightly inferior performance. Furthermore, we found that AI-based claustrum segmentation benefits from multi-view information and requires sample sizes of around 75 MRI scans in the training set. In conclusion, the developed algorithm has large potential in independent study cohorts and to facilitate MRI-based research of the human claustrum through automated segmentation. The software and models of our method are made publicly available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge