Dejan Kostyszyn
IB-U-Nets: Improving medical image segmentation tasks with 3D Inductive Biased kernels
Oct 28, 2022



Abstract:Despite the success of convolutional neural networks for 3D medical-image segmentation, the architectures currently used are still not robust enough to the protocols of different scanners, and the variety of image properties they produce. Moreover, access to large-scale datasets with annotated regions of interest is scarce, and obtaining good results is thus difficult. To overcome these challenges, we introduce IB-U-Nets, a novel architecture with inductive bias, inspired by the visual processing in vertebrates. With the 3D U-Net as the base, we add two 3D residual components to the second encoder blocks. They provide an inductive bias, helping U-Nets to segment anatomical structures from 3D images with increased robustness and accuracy. We compared IB-U-Nets with state-of-the-art 3D U-Nets on multiple modalities and organs, such as the prostate and spleen, using the same training and testing pipeline, including data processing, augmentation and cross-validation. Our results demonstrate the superior robustness and accuracy of IB-U-Nets, especially on small datasets, as is typically the case in medical-image analysis. IB-U-Nets source code and models are publicly available.
3D-OOCS: Learning Prostate Segmentation with Inductive Bias
Oct 29, 2021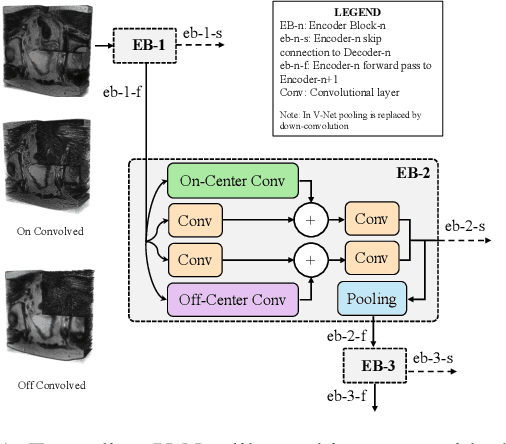


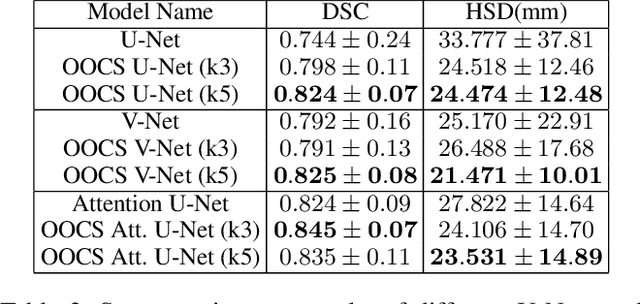
Abstract:Despite the great success of convolutional neural networks (CNN) in 3D medical image segmentation tasks, the methods currently in use are still not robust enough to the different protocols utilized by different scanners, and to the variety of image properties or artefacts they produce. To this end, we introduce OOCS-enhanced networks, a novel architecture inspired by the innate nature of visual processing in the vertebrates. With different 3D U-Net variants as the base, we add two 3D residual components to the second encoder blocks: on and off center-surround (OOCS). They generalise the ganglion pathways in the retina to a 3D setting. The use of 2D-OOCS in any standard CNN network complements the feedforward framework with sharp edge-detection inductive biases. The use of 3D-OOCS also helps 3D U-Nets to scrutinise and delineate anatomical structures present in 3D images with increased accuracy.We compared the state-of-the-art 3D U-Nets with their 3D-OOCS extensions and showed the superior accuracy and robustness of the latter in automatic prostate segmentation from 3D Magnetic Resonance Images (MRIs). For a fair comparison, we trained and tested all the investigated 3D U-Nets with the same pipeline, including automatic hyperparameter optimisation and data augmentation.
Convolutional neural network based deep-learning architecture for intraprostatic tumour contouring on PSMA PET images in patients with primary prostate cancer
Aug 07, 2020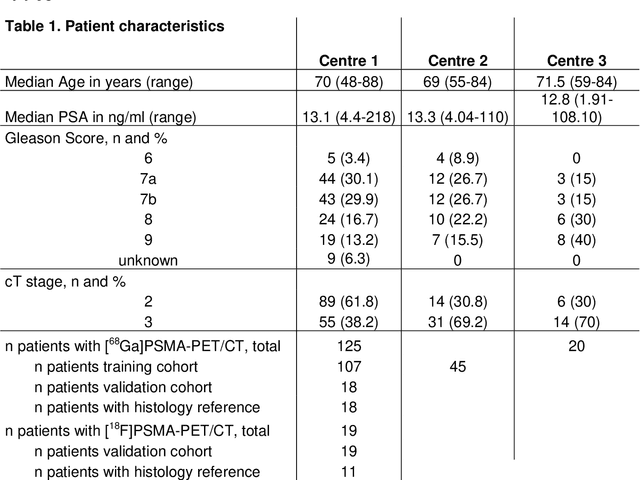
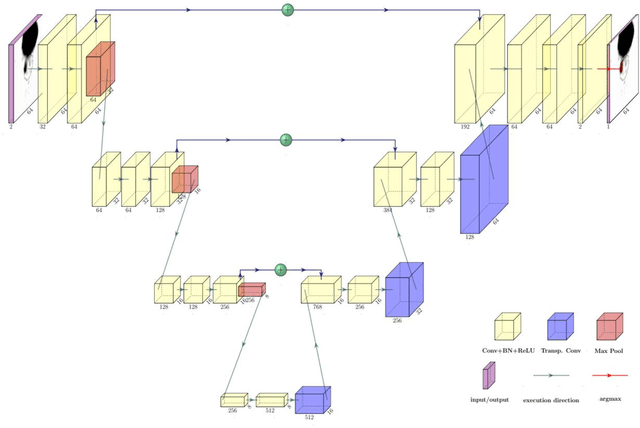
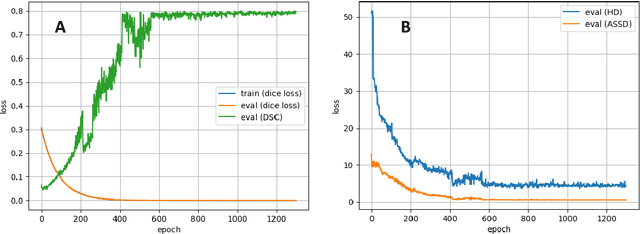
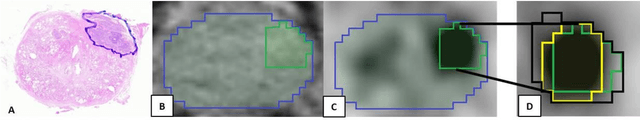
Abstract:Accurate delineation of the intraprostatic gross tumour volume (GTV) is a prerequisite for treatment approaches in patients with primary prostate cancer (PCa). Prostate-specific membrane antigen positron emission tomography (PSMA-PET) may outperform MRI in GTV detection. However, visual GTV delineation underlies interobserver heterogeneity and is time consuming. The aim of this study was to develop a convolutional neural network (CNN) for automated segmentation of intraprostatic tumour (GTV-CNN) in PSMA-PET. Methods: The CNN (3D U-Net) was trained on [68Ga]PSMA-PET images of 152 patients from two different institutions and the training labels were generated manually using a validated technique. The CNN was tested on two independent internal (cohort 1: [68Ga]PSMA-PET, n=18 and cohort 2: [18F]PSMA-PET, n=19) and one external (cohort 3: [68Ga]PSMA-PET, n=20) test-datasets. Accordance between manual contours and GTV-CNN was assessed with Dice-S{\o}rensen coefficient (DSC). Sensitivity and specificity were calculated for the two internal test-datasets by using whole-mount histology. Results: Median DSCs for cohorts 1-3 were 0.84 (range: 0.32-0.95), 0.81 (range: 0.28-0.93) and 0.83 (range: 0.32-0.93), respectively. Sensitivities and specificities for GTV-CNN were comparable with manual expert contours: 0.98 and 0.76 (cohort 1) and 1 and 0.57 (cohort 2), respectively. Computation time was around 6 seconds for a standard dataset. Conclusion: The application of a CNN for automated contouring of intraprostatic GTV in [68Ga]PSMA- and [18F]PSMA-PET images resulted in a high concordance with expert contours and in high sensitivities and specificities in comparison with histology reference. This robust, accurate and fast technique may be implemented for treatment concepts in primary PCa. The trained model and the study's source code are available in an open source repository.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge