3D-OOCS: Learning Prostate Segmentation with Inductive Bias
Paper and Code
Oct 29, 2021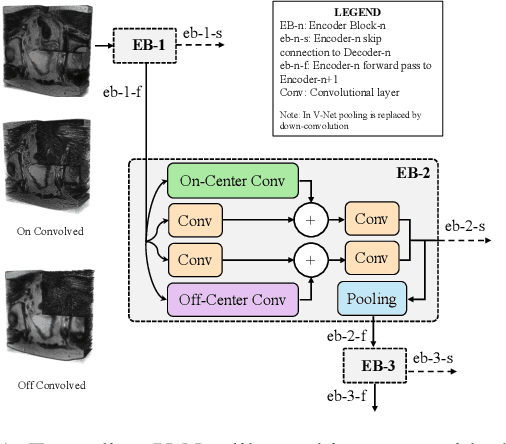


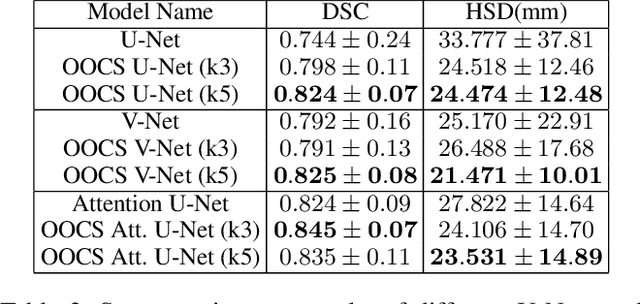
Despite the great success of convolutional neural networks (CNN) in 3D medical image segmentation tasks, the methods currently in use are still not robust enough to the different protocols utilized by different scanners, and to the variety of image properties or artefacts they produce. To this end, we introduce OOCS-enhanced networks, a novel architecture inspired by the innate nature of visual processing in the vertebrates. With different 3D U-Net variants as the base, we add two 3D residual components to the second encoder blocks: on and off center-surround (OOCS). They generalise the ganglion pathways in the retina to a 3D setting. The use of 2D-OOCS in any standard CNN network complements the feedforward framework with sharp edge-detection inductive biases. The use of 3D-OOCS also helps 3D U-Nets to scrutinise and delineate anatomical structures present in 3D images with increased accuracy.We compared the state-of-the-art 3D U-Nets with their 3D-OOCS extensions and showed the superior accuracy and robustness of the latter in automatic prostate segmentation from 3D Magnetic Resonance Images (MRIs). For a fair comparison, we trained and tested all the investigated 3D U-Nets with the same pipeline, including automatic hyperparameter optimisation and data augmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge