Darko Štern
Variational Inference and Bayesian CNNs for Uncertainty Estimation in Multi-Factorial Bone Age Prediction
Feb 25, 2020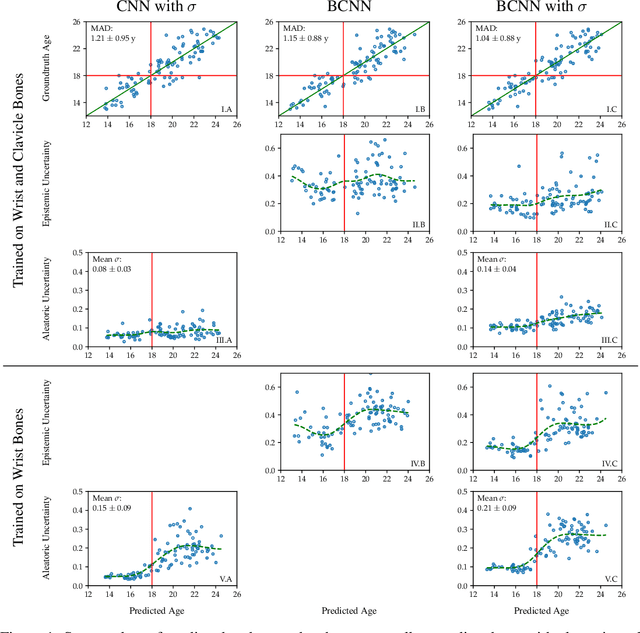
Abstract:Additionally to the extensive use in clinical medicine, biological age (BA) in legal medicine is used to assess unknown chronological age (CA) in applications where identification documents are not available. Automatic methods for age estimation proposed in the literature are predicting point estimates, which can be misleading without the quantification of predictive uncertainty. In our multi-factorial age estimation method from MRI data, we used the Variational Inference approach to estimate the uncertainty of a Bayesian CNN model. Distinguishing model uncertainty from data uncertainty, we interpreted data uncertainty as biological variation, i.e. the range of possible CA of subjects having the same BA.
Integrating Spatial Configuration into Heatmap Regression Based CNNs for Landmark Localization
Aug 02, 2019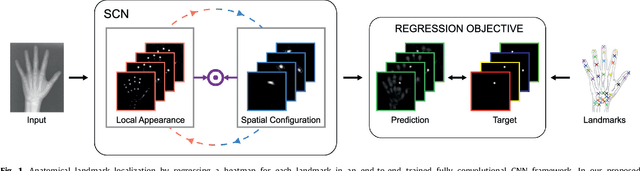
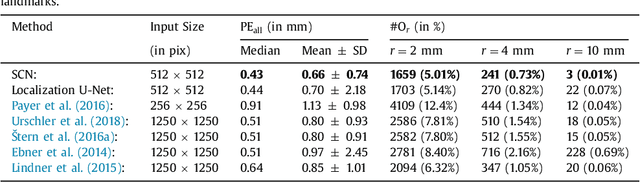
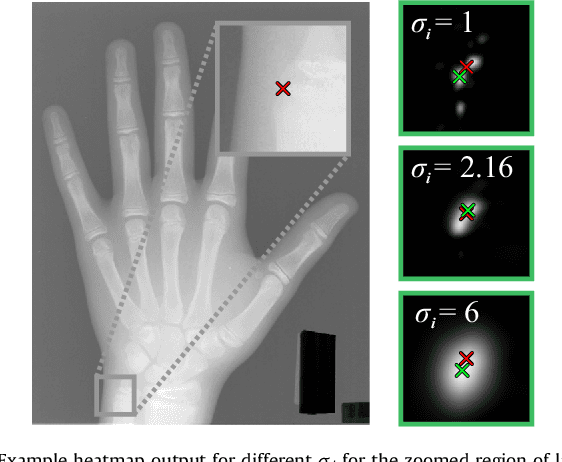
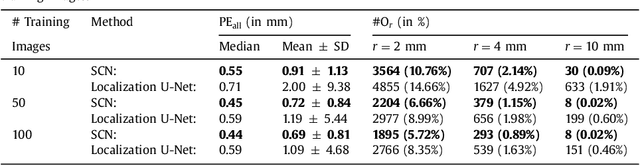
Abstract:In many medical image analysis applications, often only a limited amount of training data is available, which makes training of convolutional neural networks (CNNs) challenging. In this work on anatomical landmark localization, we propose a CNN architecture that learns to split the localization task into two simpler sub-problems, reducing the need for large training datasets. Our fully convolutional SpatialConfiguration-Net (SCN) dedicates one component to locally accurate but ambiguous candidate predictions, while the other component improves robustness to ambiguities by incorporating the spatial configuration of landmarks. In our experimental evaluation, we show that the proposed SCN outperforms related methods in terms of landmark localization error on size-limited datasets.
Instance Segmentation and Tracking with Cosine Embeddings and Recurrent Hourglass Networks
Jul 30, 2018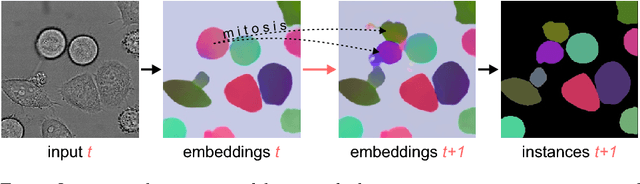
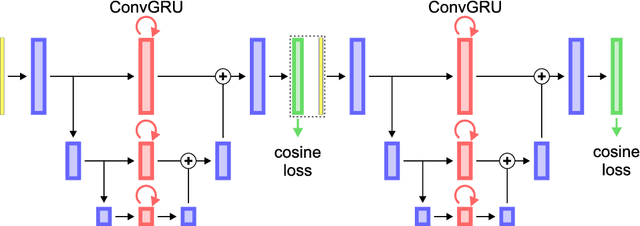
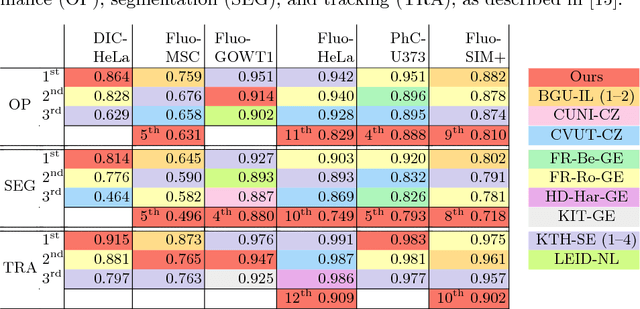
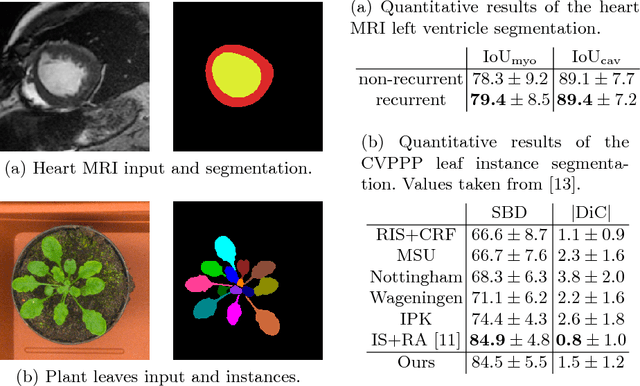
Abstract:Different to semantic segmentation, instance segmentation assigns unique labels to each individual instance of the same class. In this work, we propose a novel recurrent fully convolutional network architecture for tracking such instance segmentations over time. The network architecture incorporates convolutional gated recurrent units (ConvGRU) into a stacked hourglass network to utilize temporal video information. Furthermore, we train the network with a novel embedding loss based on cosine similarities, such that the network predicts unique embeddings for every instance throughout videos. Afterwards, these embeddings are clustered among subsequent video frames to create the final tracked instance segmentations. We evaluate the recurrent hourglass network by segmenting left ventricles in MR videos of the heart, where it outperforms a network that does not incorporate video information. Furthermore, we show applicability of the cosine embedding loss for segmenting leaf instances on still images of plants. Finally, we evaluate the framework for instance segmentation and tracking on six datasets of the ISBI celltracking challenge, where it shows state-of-the-art performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge