Cyprien Gille
Learning sparse auto-encoders for green AI image coding
Sep 09, 2022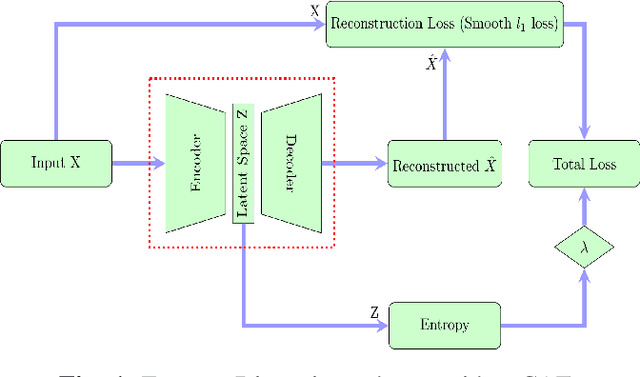
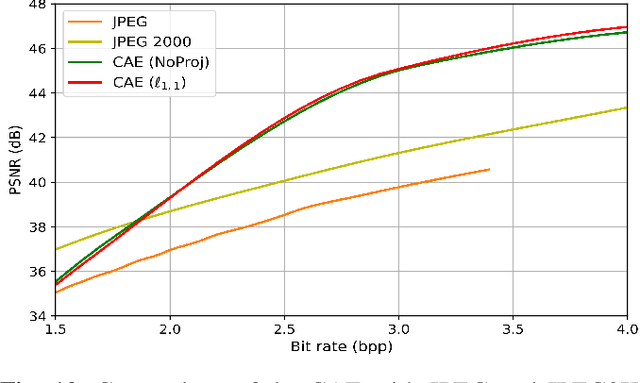
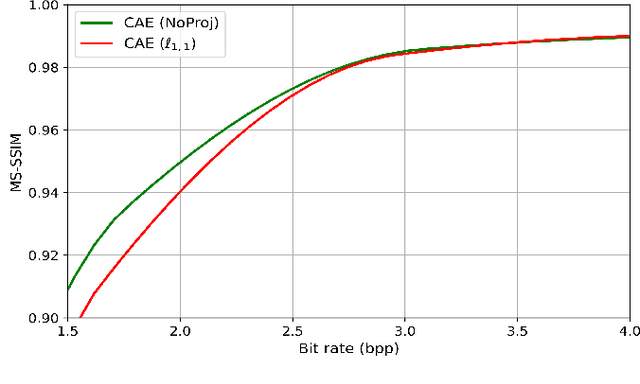
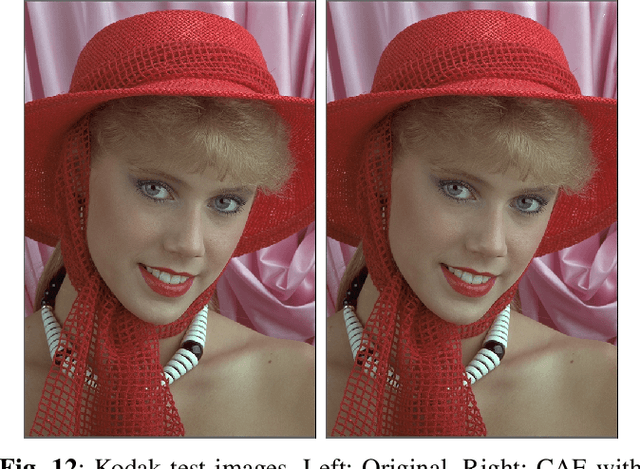
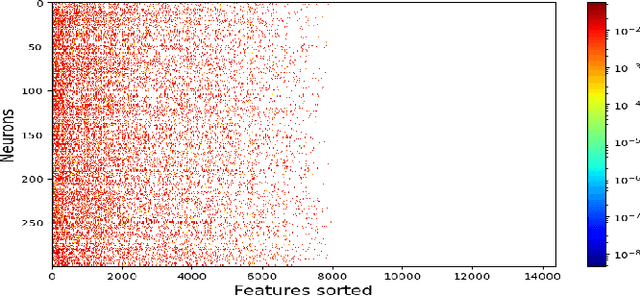
Abstract:Recently, convolutional auto-encoders (CAE) were introduced for image coding. They achieved performance improvements over the state-of-the-art JPEG2000 method. However, these performances were obtained using massive CAEs featuring a large number of parameters and whose training required heavy computational power.\\ In this paper, we address the problem of lossy image compression using a CAE with a small memory footprint and low computational power usage. In order to overcome the computational cost issue, the majority of the literature uses Lagrangian proximal regularization methods, which are time consuming themselves.\\ In this work, we propose a constrained approach and a new structured sparse learning method. We design an algorithm and test it on three constraints: the classical $\ell_1$ constraint, the $\ell_{1,\infty}$ and the new $\ell_{1,1}$ constraint. Experimental results show that the $\ell_{1,1}$ constraint provides the best structured sparsity, resulting in a high reduction of memory and computational cost, with similar rate-distortion performance as with dense networks.
Semi-supervised classification using a supervised autoencoder for biomedical applications
Aug 22, 2022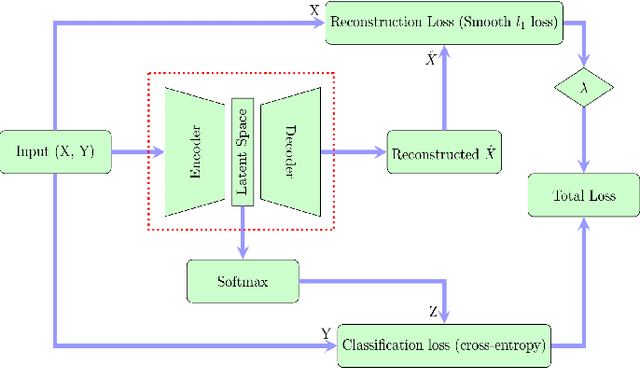
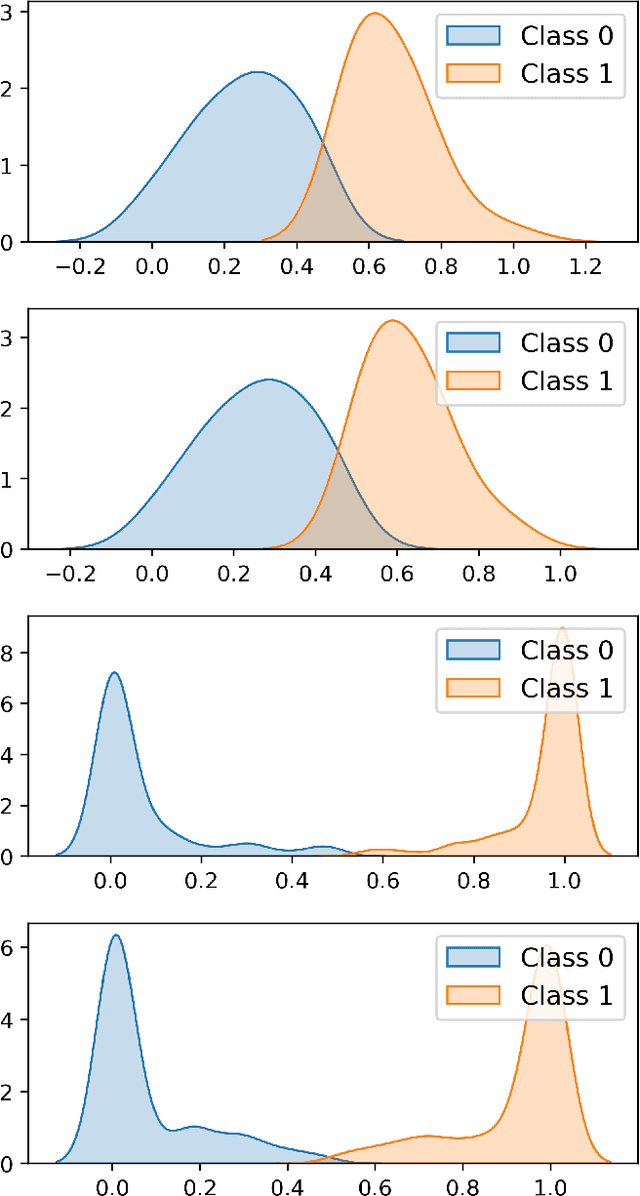
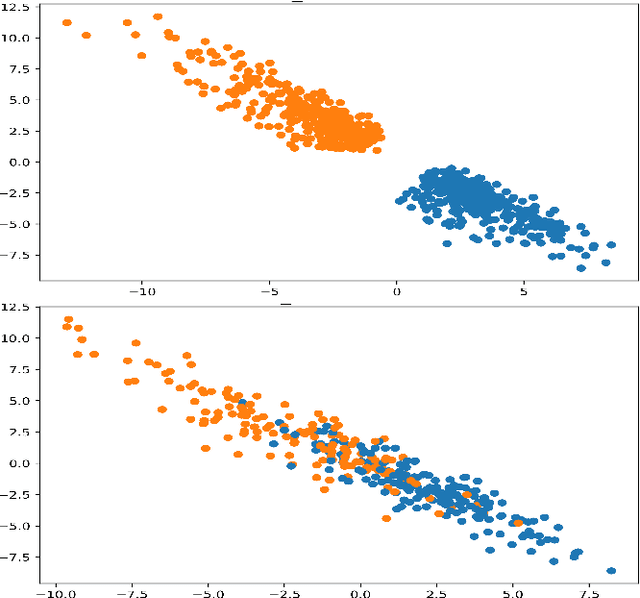
Abstract:In this paper we present a new approach to solve semi-supervised classification tasks for biomedical applications, involving a supervised autoencoder network. We create a network architecture that encodes labels into the latent space of an autoencoder, and define a global criterion combining classification and reconstruction losses. We train the Semi-Supervised AutoEncoder (SSAE) on labelled data using a double descent algorithm. Then, we classify unlabelled samples using the learned network thanks to a softmax classifier applied to the latent space which provides a classification confidence score for each class. We implemented our SSAE method using the PyTorch framework for the model, optimizer, schedulers, and loss functions. We compare our semi-supervised autoencoder method (SSAE) with classical semi-supervised methods such as Label Propagation and Label Spreading, and with a Fully Connected Neural Network (FCNN). Experiments show that the SSAE outperforms Label Propagation and Spreading and the Fully Connected Neural Network both on a synthetic dataset and on two real-world biological datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge