Semi-supervised classification using a supervised autoencoder for biomedical applications
Paper and Code
Aug 22, 2022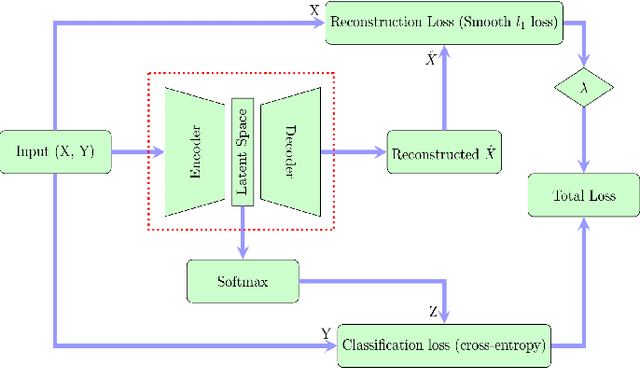
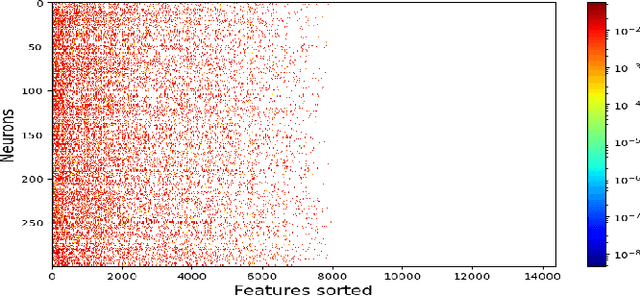
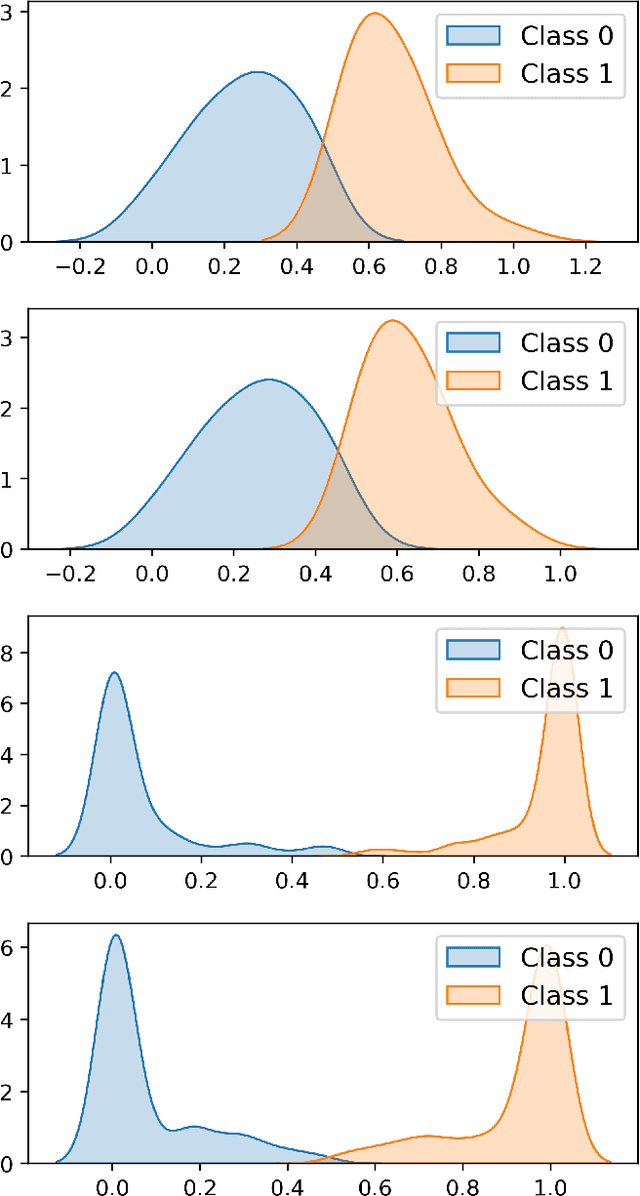
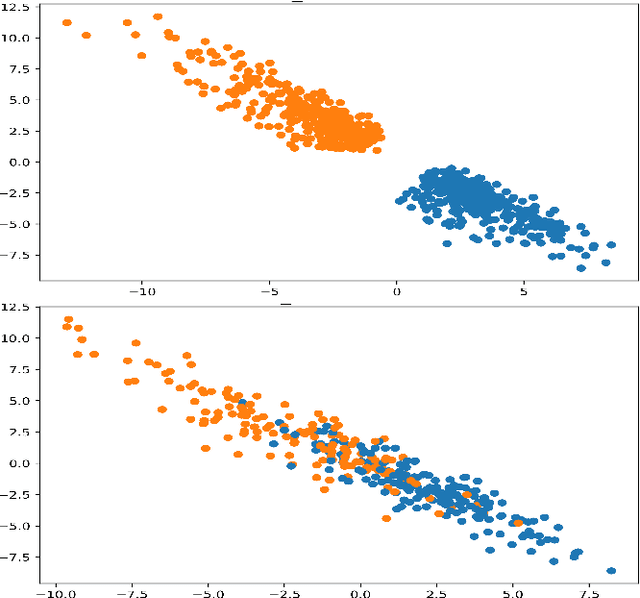
In this paper we present a new approach to solve semi-supervised classification tasks for biomedical applications, involving a supervised autoencoder network. We create a network architecture that encodes labels into the latent space of an autoencoder, and define a global criterion combining classification and reconstruction losses. We train the Semi-Supervised AutoEncoder (SSAE) on labelled data using a double descent algorithm. Then, we classify unlabelled samples using the learned network thanks to a softmax classifier applied to the latent space which provides a classification confidence score for each class. We implemented our SSAE method using the PyTorch framework for the model, optimizer, schedulers, and loss functions. We compare our semi-supervised autoencoder method (SSAE) with classical semi-supervised methods such as Label Propagation and Label Spreading, and with a Fully Connected Neural Network (FCNN). Experiments show that the SSAE outperforms Label Propagation and Spreading and the Fully Connected Neural Network both on a synthetic dataset and on two real-world biological datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge