Colin Targonski
Cellular State Transformations using Generative Adversarial Networks
Jun 28, 2019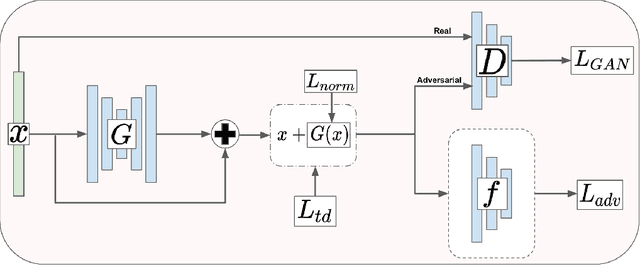
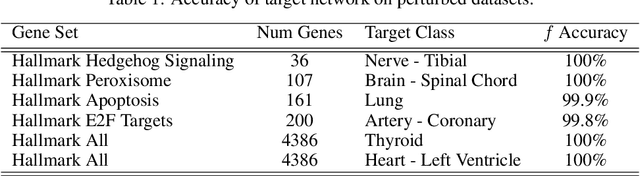
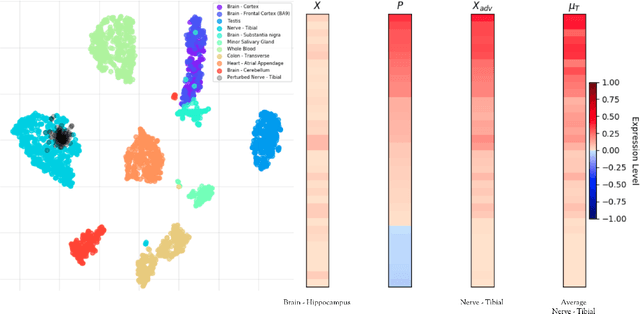
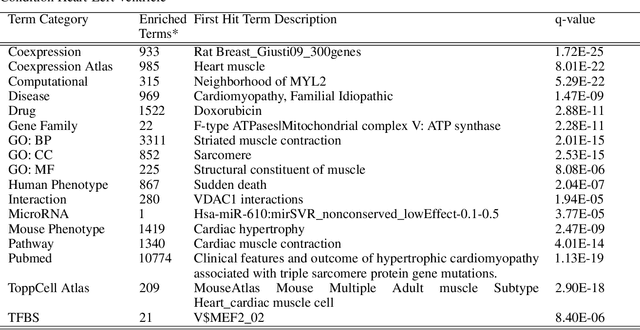
Abstract:We introduce a novel method to unite deep learning with biology by which generative adversarial networks (GANs) generate transcriptome perturbations and reveal condition-defining gene expression patterns. We find that a generator conditioned to perturb any input gene expression profile simulates a realistic transition between source and target RNA expression states. The perturbed samples follow a similar distribution to original samples from the dataset, also suggesting these are biologically meaningful perturbations. Finally, we show that it is possible to identify the genes most positively and negatively perturbed by the generator and that the enriched biological function of the perturbed genes are realistic. We call the framework the Transcriptome State Perturbation Generator (TSPG), which is open source software available at https://github.com/ctargon/TSPG.
Generalized Value Iteration Networks: Life Beyond Lattices
Oct 26, 2017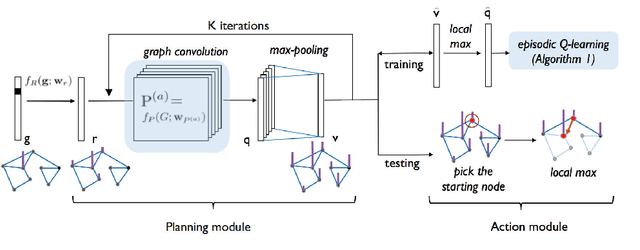
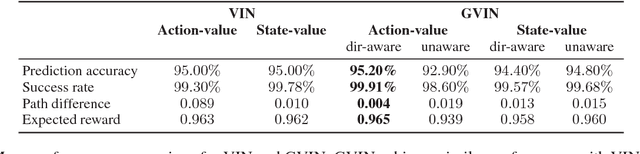
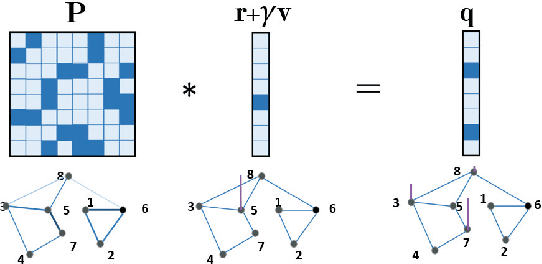
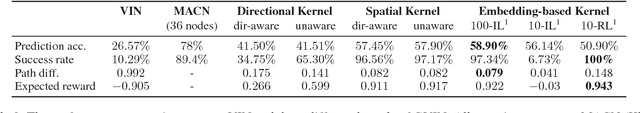
Abstract:In this paper, we introduce a generalized value iteration network (GVIN), which is an end-to-end neural network planning module. GVIN emulates the value iteration algorithm by using a novel graph convolution operator, which enables GVIN to learn and plan on irregular spatial graphs. We propose three novel differentiable kernels as graph convolution operators and show that the embedding based kernel achieves the best performance. We further propose episodic Q-learning, an improvement upon traditional n-step Q-learning that stabilizes training for networks that contain a planning module. Lastly, we evaluate GVIN on planning problems in 2D mazes, irregular graphs, and real-world street networks, showing that GVIN generalizes well for both arbitrary graphs and unseen graphs of larger scale and outperforms a naive generalization of VIN (discretizing a spatial graph into a 2D image).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge