Cláudio R. Jung
Cell Tracking-by-detection using Elliptical Bounding Boxes
Oct 11, 2023



Abstract:Cell detection and tracking are paramount for bio-analysis. Recent approaches rely on the tracking-by-model evolution paradigm, which usually consists of training end-to-end deep learning models to detect and track the cells on the frames with promising results. However, such methods require extensive amounts of annotated data, which is time-consuming to obtain and often requires specialized annotators. This work proposes a new approach based on the classical tracking-by-detection paradigm that alleviates the requirement of annotated data. More precisely, it approximates the cell shapes as oriented ellipses and then uses generic-purpose oriented object detectors to identify the cells in each frame. We then rely on a global data association algorithm that explores temporal cell similarity using probability distance metrics, considering that the ellipses relate to two-dimensional Gaussian distributions. Our results show that our method can achieve detection and tracking results competitively with state-of-the-art techniques that require considerably more extensive data annotation. Our code is available at: https://github.com/LucasKirsten/Deep-Cell-Tracking-EBB.
Superpixel Image Classification with Graph Attention Networks
Feb 13, 2020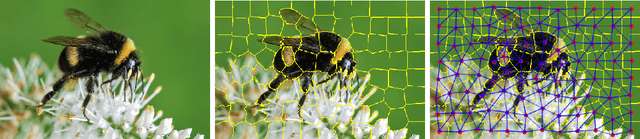
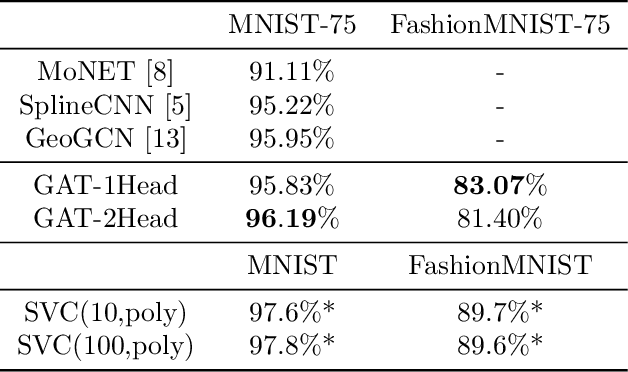
Abstract:This document reports the use of Graph Attention Networks for classifying oversegmented images, as well as a general procedure for generating oversegmented versions of image-based datasets. The code and learnt models for/from the experiments are available on github. The experiments were ran from June 2019 until December 2019. We obtained better results than the baseline models that uses geometric distance-based attention by using instead self attention, in a more sparsely connected graph network.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge