Christopher G. Knight
Monotonicity of Fitness Landscapes and Mutation Rate Control
Sep 04, 2012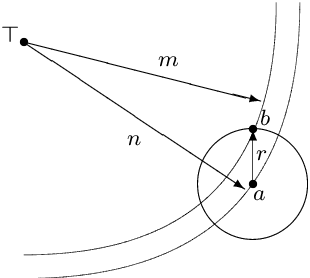
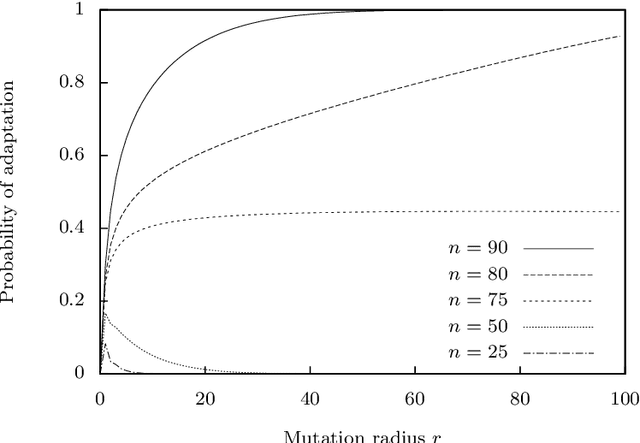
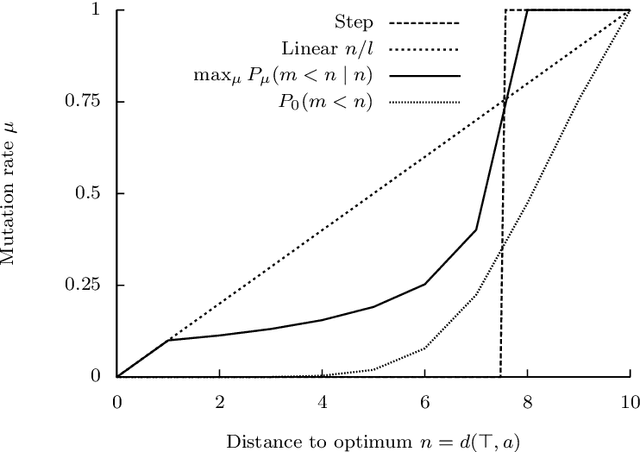
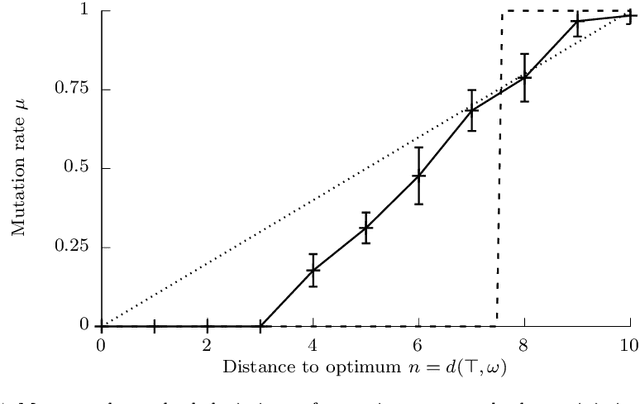
Abstract:The typical view in evolutionary biology is that mutation rates are minimised. Contrary to that view, studies in combinatorial optimisation and search have shown a clear advantage of using variable mutation rates as a control parameter to optimise the performance of evolutionary algorithms. Ronald Fisher's work is the basis of much biological theory in this area. He used Euclidean geometry of continuous, infinite phenotypic spaces to study the relation between mutation size and expected fitness of the offspring. Here we develop a general theory of optimal mutation rate control that is based on the alternative geometry of discrete and finite spaces of DNA sequences. We define the monotonic properties of fitness landscapes, which allows us to relate fitness to the topology of genotypes and mutation size. First, we consider the case of a perfectly monotonic fitness landscape, in which the optimal mutation rate control functions can be derived exactly or approximately depending on additional constraints of the problem. Then we consider the general case of non-monotonic landscapes. We use the ideas of local and weak monotonicity to show that optimal mutation rate control functions exist in any such landscape and that they resemble control functions in a monotonic landscape at least in some neighbourhood of a fitness maximum. Generally, optimal mutation rates increase when fitness decreases, and the increase of mutation rate is more rapid in landscapes that are less monotonic (more rugged). We demonstrate these relationships by obtaining and analysing approximately optimal mutation rate control functions in 115 complete landscapes of binding scores between DNA sequences and transcription factors. We discuss the relevance of these findings to living organisms, including the phenomenon of stress-induced mutagenesis.
Ancestral Inference from Functional Data: Statistical Methods and Numerical Examples
Aug 02, 2012
Abstract:Many biological characteristics of evolutionary interest are not scalar variables but continuous functions. Here we use phylogenetic Gaussian process regression to model the evolution of simulated function-valued traits. Given function-valued data only from the tips of an evolutionary tree and utilising independent principal component analysis (IPCA) as a method for dimension reduction, we construct distributional estimates of ancestral function-valued traits, and estimate parameters describing their evolutionary dynamics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge