Christine Picard
Fine-Grained Zero-Shot Learning with DNA as Side Information
Sep 29, 2021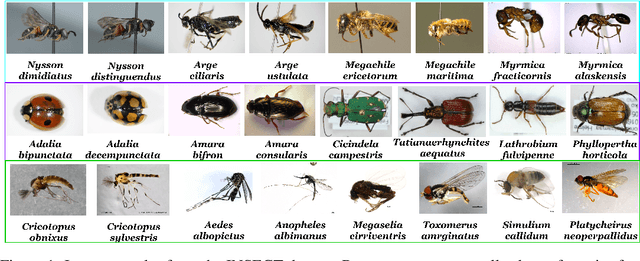
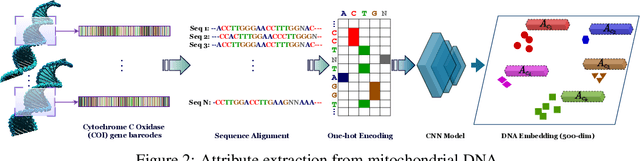
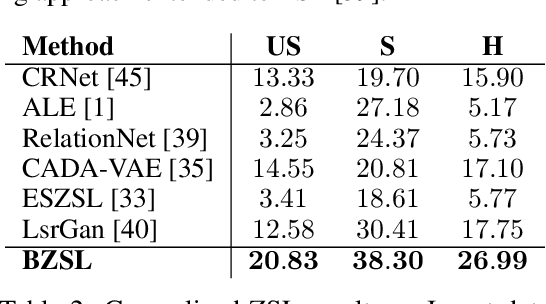
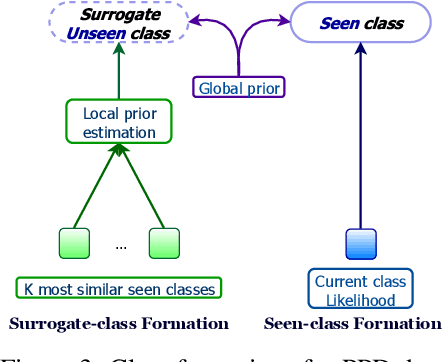
Abstract:Fine-grained zero-shot learning task requires some form of side-information to transfer discriminative information from seen to unseen classes. As manually annotated visual attributes are extremely costly and often impractical to obtain for a large number of classes, in this study we use DNA as side information for the first time for fine-grained zero-shot classification of species. Mitochondrial DNA plays an important role as a genetic marker in evolutionary biology and has been used to achieve near-perfect accuracy in the species classification of living organisms. We implement a simple hierarchical Bayesian model that uses DNA information to establish the hierarchy in the image space and employs local priors to define surrogate classes for unseen ones. On the benchmark CUB dataset, we show that DNA can be equally promising yet in general a more accessible alternative than word vectors as a side information. This is especially important as obtaining robust word representations for fine-grained species names is not a practicable goal when information about these species in free-form text is limited. On a newly compiled fine-grained insect dataset that uses DNA information from over a thousand species, we show that the Bayesian approach outperforms state-of-the-art by a wide margin.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge