Christine Devalland
HNFC
A new methodology to predict the oncotype scores based on clinico-pathological data with similar tumor profiles
Mar 13, 2023Abstract:Introduction: The Oncotype DX (ODX) test is a commercially available molecular test for breast cancer assay that provides prognostic and predictive breast cancer recurrence information for hormone positive, HER2-negative patients. The aim of this study is to propose a novel methodology to assist physicians in their decision-making. Methods: A retrospective study between 2012 and 2020 with 333 cases that underwent an ODX assay from three hospitals in Bourgogne Franche-Comt{\'e} was conducted. Clinical and pathological reports were used to collect the data. A methodology based on distributional random forest was developed using 9 clinico-pathological characteristics. This methodology can be used particularly to identify the patients of the training cohort that share similarities with the new patient and to predict an estimate of the distribution of the ODX score. Results: The mean age of participants id 56.9 years old. We have correctly classified 92% of patients in low risk and 40.2% of patients in high risk. The overall accuracy is 79.3%. The proportion of low risk correct predicted value (PPV) is 82%. The percentage of high risk correct predicted value (NPV) is approximately 62.3%. The F1-score and the Area Under Curve (AUC) are of 0.87 and 0.759, respectively. Conclusion: The proposed methodology makes it possible to predict the distribution of the ODX score for a patient and provides an explanation of the predicted score. The use of the methodology with the pathologist's expertise on the different histological and immunohistochemical characteristics has a clinical impact to help oncologist in decision-making regarding breast cancer therapy.
A deep learning pipeline for breast cancer ki-67 proliferation index scoring
Mar 14, 2022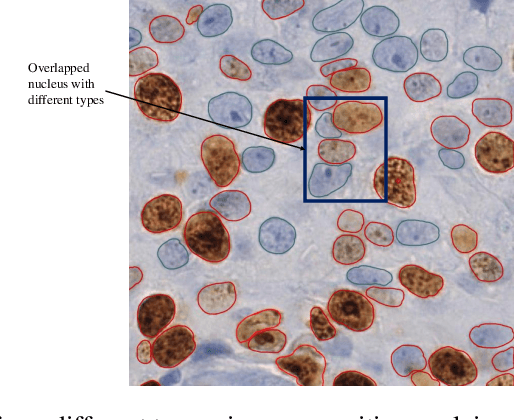
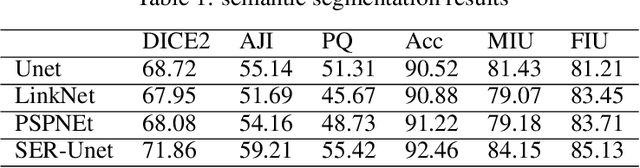
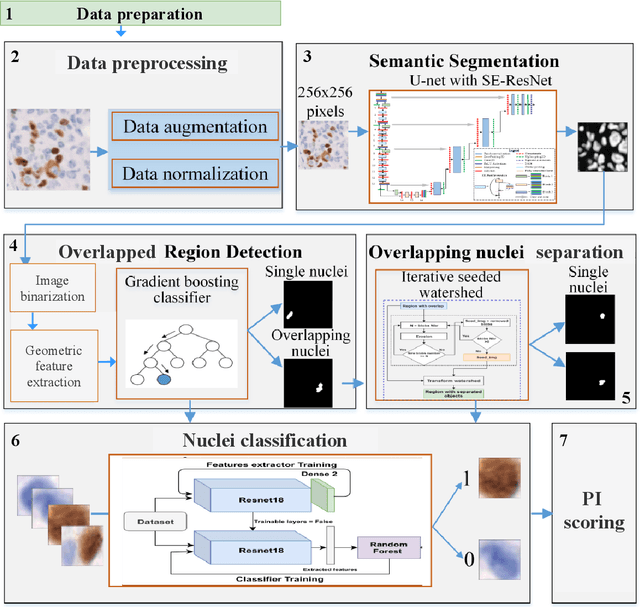
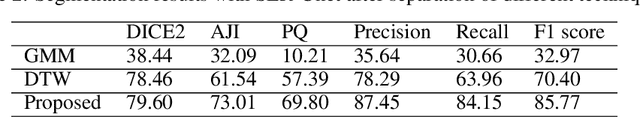
Abstract:The Ki-67 proliferation index is an essential biomarker that helps pathologists to diagnose and select appropriate treatments. However, automatic evaluation of Ki-67 is difficult due to nuclei overlapping and complex variations in their properties. This paper proposes an integrated pipeline for accurate automatic counting of Ki-67, where the impact of nuclei separation techniques is highlighted. First, semantic segmentation is performed by combining the Squeez and Excitation Resnet and Unet algorithms to extract nuclei from the background. The extracted nuclei are then divided into overlapped and non-overlapped regions based on eight geometric and statistical features. A marker-based Watershed algorithm is subsequently proposed and applied only to the overlapped regions to separate nuclei. Finally, deep features are extracted from each nucleus patch using Resnet18 and classified into positive or negative by a random forest classifier. The proposed pipeline's performance is validated on a dataset from the Department of Pathology at H\^opital Nord Franche-Comt\'e hospital.
Computer Aided Diagnosis for Spitzoid lesions classification using Artificial Intelligence techniques
Mar 10, 2020
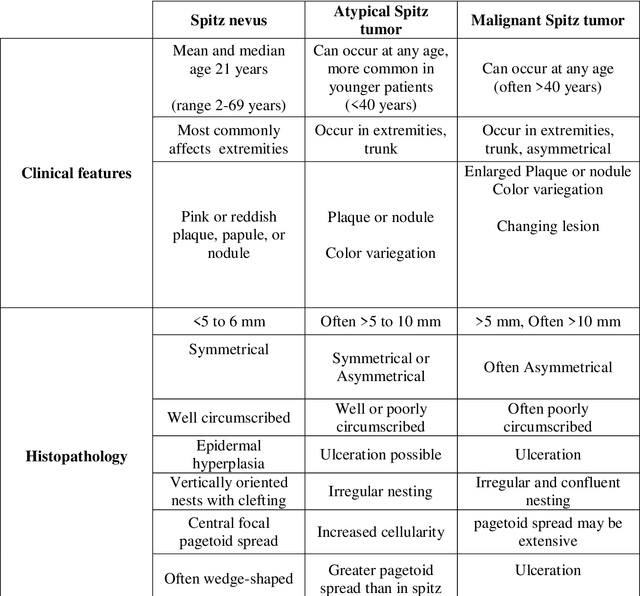
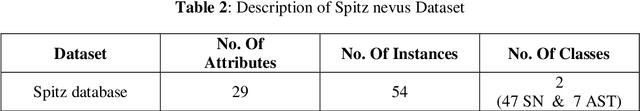

Abstract:Spitzoid lesions may be largely categorized into Spitz Nevus, Atypical Spitz Tumors, and Spitz Melanomas. Classifying a lesion precisely as Atypical Spitz Tumors or AST is challenging and often requires the integration of clinical, histological, and immunohistochemical features to differentiate AST from regular Spitz nevus and malignant Spitz melanomas. Specifically, this paper aims to test several artificial intelligence techniques so as to build a computer aided diagnosis system. A proposed three-phase approach is being implemented. In Phase I, collected data are preprocessed with an effective Synthetic Minority Oversampling TEchnique or SMOTE-based method being implemented to treat the imbalance data problem. Then, a feature selection mechanism using genetic algorithm (GA) is applied in Phase II. Finally, in Phase III, a ten-fold cross-validation method is used to compare the performance of seven machine-learning algorithms for classification. Results obtained with SMOTE-Multilayer Perceptron with GA-based 14 features show the highest classification accuracy (0.98), a sensitivity of 0.99, and a specificity of 0.98, outperforming other Spitzoid lesions classification algorithms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge