A deep learning pipeline for breast cancer ki-67 proliferation index scoring
Paper and Code
Mar 14, 2022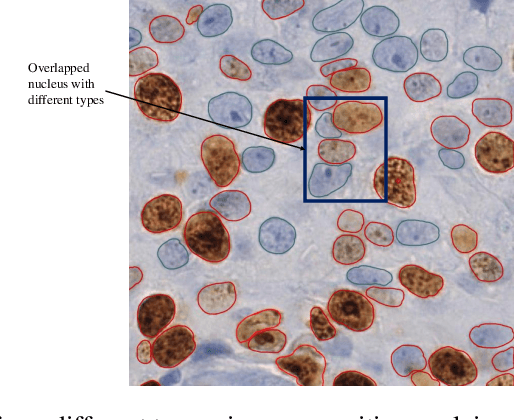
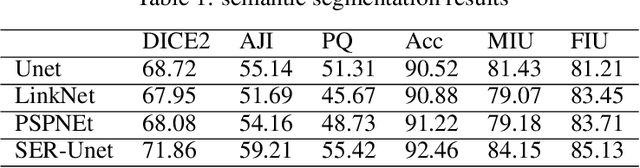
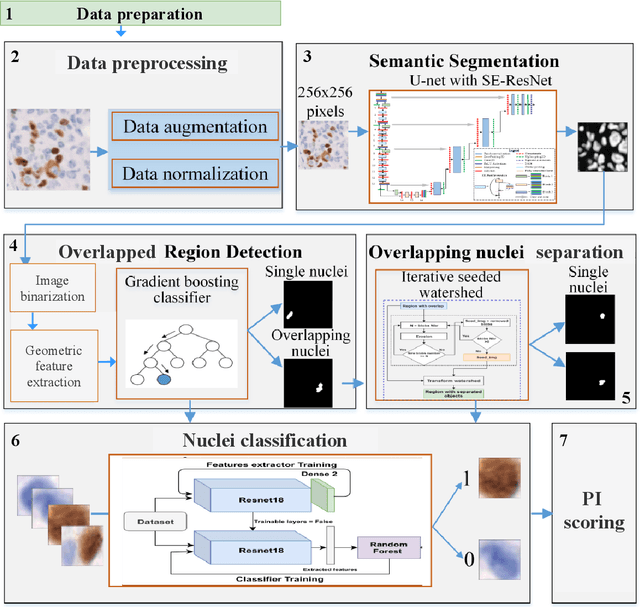
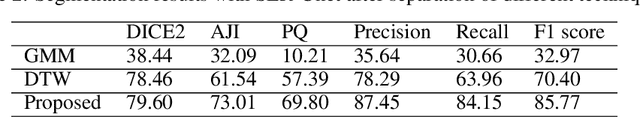
The Ki-67 proliferation index is an essential biomarker that helps pathologists to diagnose and select appropriate treatments. However, automatic evaluation of Ki-67 is difficult due to nuclei overlapping and complex variations in their properties. This paper proposes an integrated pipeline for accurate automatic counting of Ki-67, where the impact of nuclei separation techniques is highlighted. First, semantic segmentation is performed by combining the Squeez and Excitation Resnet and Unet algorithms to extract nuclei from the background. The extracted nuclei are then divided into overlapped and non-overlapped regions based on eight geometric and statistical features. A marker-based Watershed algorithm is subsequently proposed and applied only to the overlapped regions to separate nuclei. Finally, deep features are extracted from each nucleus patch using Resnet18 and classified into positive or negative by a random forest classifier. The proposed pipeline's performance is validated on a dataset from the Department of Pathology at H\^opital Nord Franche-Comt\'e hospital.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge