Changfa Shi
Bend-Net: Bending Loss Regularized Multitask Learning Network for Nuclei Segmentation in Histopathology Images
Sep 30, 2021



Abstract:Separating overlapped nuclei is a major challenge in histopathology image analysis. Recently published approaches have achieved promising overall performance on nuclei segmentation; however, their performance on separating overlapped nuclei is quite limited. To address the issue, we propose a novel multitask learning network with a bending loss regularizer to separate overlapped nuclei accurately. The newly proposed multitask learning architecture enhances the generalization by learning shared representation from three tasks: instance segmentation, nuclei distance map prediction, and overlapped nuclei distance map prediction. The proposed bending loss defines high penalties to concave contour points with large curvatures, and applies small penalties to convex contour points with small curvatures. Minimizing the bending loss avoids generating contours that encompass multiple nuclei. In addition, two new quantitative metrics, Aggregated Jaccard Index of overlapped nuclei (AJIO) and Accuracy of overlapped nuclei (ACCO), are designed for the evaluation of overlapped nuclei segmentation. We validate the proposed approach on the CoNSeP and MoNuSegv1 datasets using seven quantitative metrics: Aggregate Jaccard Index, Dice, Segmentation Quality, Recognition Quality, Panoptic Quality, AJIO, and ACCO. Extensive experiments demonstrate that the proposed Bend-Net outperforms eight state-of-the-art approaches.
SAR-U-Net: squeeze-and-excitation block and atrous spatial pyramid pooling based residual U-Net for automatic liver CT segmentation
Mar 11, 2021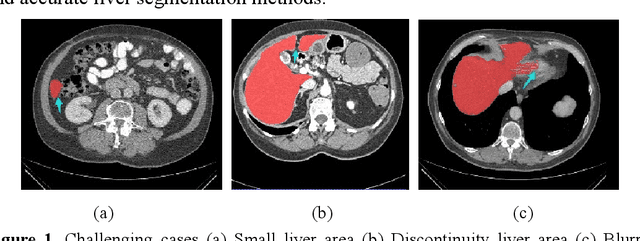
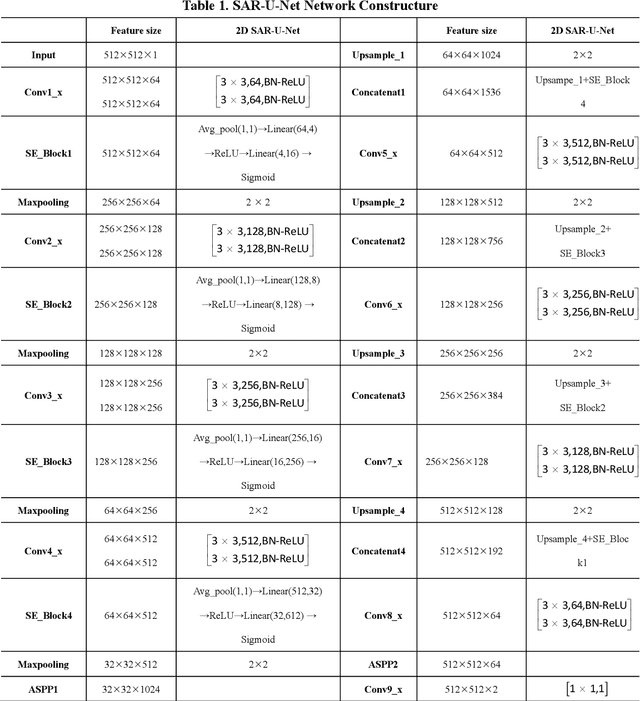
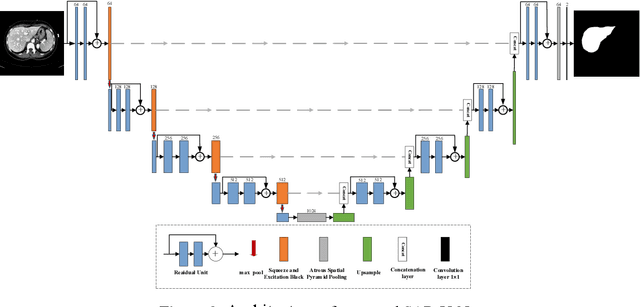
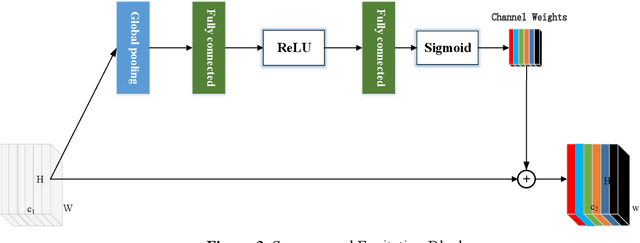
Abstract:Background and objective: In this paper, a modified U-Net based framework is presented, which leverages techniques from Squeeze-and-Excitation (SE) block, Atrous Spatial Pyramid Pooling (ASPP) and residual learning for accurate and robust liver CT segmentation, and the effectiveness of the proposed method was tested on two public datasets LiTS17 and SLiver07. Methods: A new network architecture called SAR-U-Net was designed. Firstly, the SE block is introduced to adaptively extract image features after each convolution in the U-Net encoder, while suppressing irrelevant regions, and highlighting features of specific segmentation task; Secondly, ASPP was employed to replace the transition layer and the output layer, and acquire multi-scale image information via different receptive fields. Thirdly, to alleviate the degradation problem, the traditional convolution block was replaced with the residual block and thus prompt the network to gain accuracy from considerably increased depth. Results: In the LiTS17 experiment, the mean values of Dice, VOE, RVD, ASD and MSD were 95.71, 9.52, -0.84, 1.54 and 29.14, respectively. Compared with other closely related 2D-based models, the proposed method achieved the highest accuracy. In the experiment of the SLiver07, the mean values of Dice, VOE, RVD, ASD and MSD were 97.31, 5.37, -1.08, 1.85 and 27.45, respectively. Compared with other closely related models, the proposed method achieved the highest segmentation accuracy except for the RVD. Conclusion: The proposed model enables a great improvement on the accuracy compared to 2D-based models, and its robustness in circumvent challenging problems, such as small liver regions, discontinuous liver regions, and fuzzy liver boundaries, is also well demonstrated and validated.
Multi-Slice Low-Rank Tensor Decomposition Based Multi-Atlas Segmentation: Application to Automatic Pathological Liver CT Segmentation
Feb 24, 2021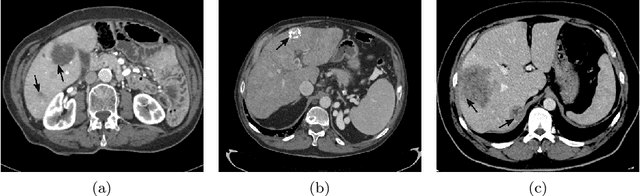
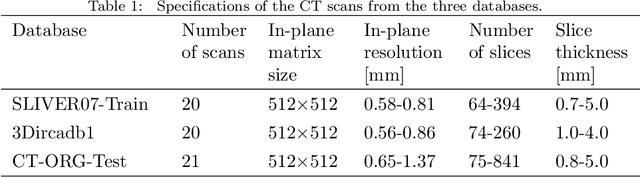
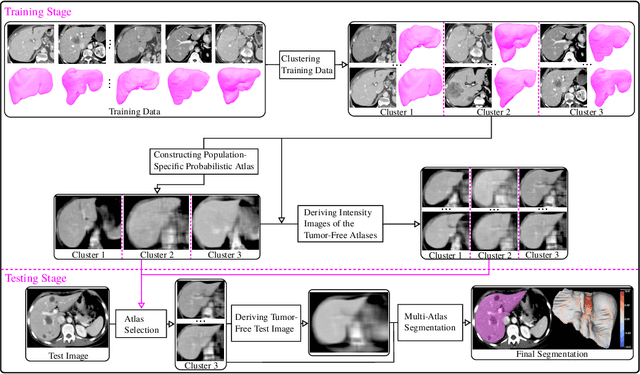
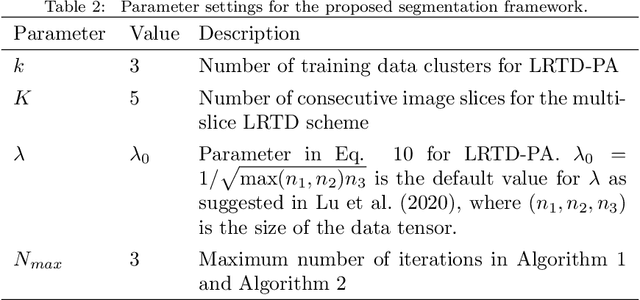
Abstract:Liver segmentation from abdominal CT images is an essential step for liver cancer computer-aided diagnosis and surgical planning. However, both the accuracy and robustness of existing liver segmentation methods cannot meet the requirements of clinical applications. In particular, for the common clinical cases where the liver tissue contains major pathology, current segmentation methods show poor performance. In this paper, we propose a novel low-rank tensor decomposition (LRTD) based multi-atlas segmentation (MAS) framework that achieves accurate and robust pathological liver segmentation of CT images. Firstly, we propose a multi-slice LRTD scheme to recover the underlying low-rank structure embedded in 3D medical images. It performs the LRTD on small image segments consisting of multiple consecutive image slices. Then, we present an LRTD-based atlas construction method to generate tumor-free liver atlases that mitigates the performance degradation of liver segmentation due to the presence of tumors. Finally, we introduce an LRTD-based MAS algorithm to derive patient-specific liver atlases for each test image, and to achieve accurate pairwise image registration and label propagation. Extensive experiments on three public databases of pathological liver cases validate the effectiveness of the proposed method. Both qualitative and quantitative results demonstrate that, in the presence of major pathology, the proposed method is more accurate and robust than state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge