Aleksandar Vakanski
Decentralized Distributed Proximal Policy Optimization (DD-PPO) for High Performance Computing Scheduling on Multi-User Systems
May 06, 2025Abstract:Resource allocation in High Performance Computing (HPC) environments presents a complex and multifaceted challenge for job scheduling algorithms. Beyond the efficient allocation of system resources, schedulers must account for and optimize multiple performance metrics, including job wait time and system utilization. While traditional rule-based scheduling algorithms dominate the current deployments of HPC systems, the increasing heterogeneity and scale of those systems is expected to challenge the efficiency and flexibility of those algorithms in minimizing job wait time and maximizing utilization. Recent research efforts have focused on leveraging advancements in Reinforcement Learning (RL) to develop more adaptable and intelligent scheduling strategies. Recent RL-based scheduling approaches have explored a range of algorithms, from Deep Q-Networks (DQN) to Proximal Policy Optimization (PPO), and more recently, hybrid methods that integrate Graph Neural Networks with RL techniques. However, a common limitation across these methods is their reliance on relatively small datasets, and these methods face scalability issues when using large datasets. This study introduces a novel RL-based scheduler utilizing the Decentralized Distributed Proximal Policy Optimization (DD-PPO) algorithm, which supports large-scale distributed training across multiple workers without requiring parameter synchronization at every step. By eliminating reliance on centralized updates to a shared policy, the DD-PPO scheduler enhances scalability, training efficiency, and sample utilization. The validation dataset leveraged over 11.5 million real HPC job traces for comparing DD-PPO performance between traditional and advanced scheduling approaches, and the experimental results demonstrate improved scheduling performance in comparison to both rule-based schedulers and existing RL-based scheduling algorithms.
Graph Kolmogorov-Arnold Networks for Multi-Cancer Classification and Biomarker Identification, An Interpretable Multi-Omics Approach
Mar 29, 2025Abstract:The integration of multi-omics data presents a major challenge in precision medicine, requiring advanced computational methods for accurate disease classification and biological interpretation. This study introduces the Multi-Omics Graph Kolmogorov-Arnold Network (MOGKAN), a deep learning model that integrates messenger RNA, micro RNA sequences, and DNA methylation data with Protein-Protein Interaction (PPI) networks for accurate and interpretable cancer classification across 31 cancer types. MOGKAN employs a hybrid approach combining differential expression with DESeq2, Linear Models for Microarray (LIMMA), and Least Absolute Shrinkage and Selection Operator (LASSO) regression to reduce multi-omics data dimensionality while preserving relevant biological features. The model architecture is based on the Kolmogorov-Arnold theorem principle, using trainable univariate functions to enhance interpretability and feature analysis. MOGKAN achieves classification accuracy of 96.28 percent and demonstrates low experimental variability with a standard deviation that is reduced by 1.58 to 7.30 percents compared to Convolutional Neural Networks (CNNs) and Graph Neural Networks (GNNs). The biomarkers identified by MOGKAN have been validated as cancer-related markers through Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis. The proposed model presents an ability to uncover molecular oncogenesis mechanisms by detecting phosphoinositide-binding substances and regulating sphingolipid cellular processes. By integrating multi-omics data with graph-based deep learning, our proposed approach demonstrates superior predictive performance and interpretability that has the potential to enhance the translation of complex multi-omics data into clinically actionable cancer diagnostics.
Predictive Modeling and Uncertainty Quantification of Fatigue Life in Metal Alloys using Machine Learning
Jan 25, 2025
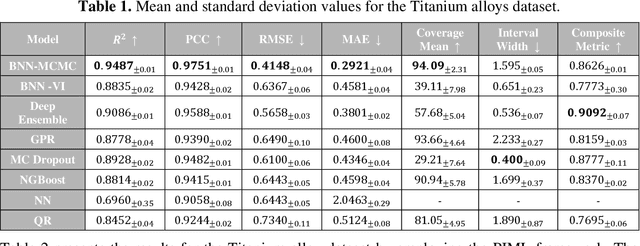
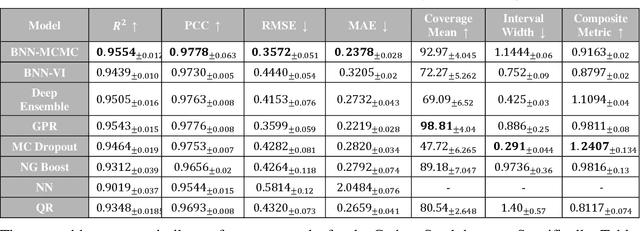
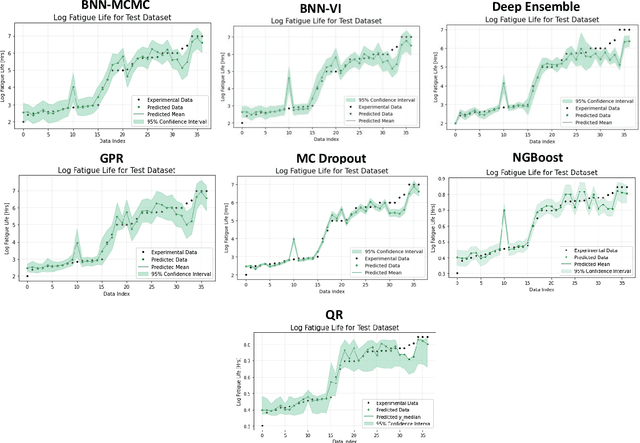
Abstract:Recent advancements in machine learning-based methods have demonstrated great potential for improved property prediction in material science. However, reliable estimation of the confidence intervals for the predicted values remains a challenge, due to the inherent complexities in material modeling. This study introduces a novel approach for uncertainty quantification in fatigue life prediction of metal materials based on integrating knowledge from physics-based fatigue life models and machine learning models. The proposed approach employs physics-based input features estimated using the Basquin fatigue model to augment the experimentally collected data of fatigue life. Furthermore, a physics-informed loss function that enforces boundary constraints for the estimated fatigue life of considered materials is introduced for the neural network models. Experimental validation on datasets comprising collected data from fatigue life tests for Titanium alloys and Carbon steel alloys demonstrates the effectiveness of the proposed approach. The synergy between physics-based models and data-driven models enhances the consistency in predicted values and improves uncertainty interval estimates.
GCSAM: Gradient Centralized Sharpness Aware Minimization
Jan 20, 2025Abstract:The generalization performance of deep neural networks (DNNs) is a critical factor in achieving robust model behavior on unseen data. Recent studies have highlighted the importance of sharpness-based measures in promoting generalization by encouraging convergence to flatter minima. Among these approaches, Sharpness-Aware Minimization (SAM) has emerged as an effective optimization technique for reducing the sharpness of the loss landscape, thereby improving generalization. However, SAM's computational overhead and sensitivity to noisy gradients limit its scalability and efficiency. To address these challenges, we propose Gradient-Centralized Sharpness-Aware Minimization (GCSAM), which incorporates Gradient Centralization (GC) to stabilize gradients and accelerate convergence. GCSAM normalizes gradients before the ascent step, reducing noise and variance, and improving stability during training. Our evaluations indicate that GCSAM consistently outperforms SAM and the Adam optimizer in terms of generalization and computational efficiency. These findings demonstrate GCSAM's effectiveness across diverse domains, including general and medical imaging tasks.
Comparative Analysis of Multi-Omics Integration Using Advanced Graph Neural Networks for Cancer Classification
Oct 05, 2024



Abstract:Multi-omics data is increasingly being utilized to advance computational methods for cancer classification. However, multi-omics data integration poses significant challenges due to the high dimensionality, data complexity, and distinct characteristics of various omics types. This study addresses these challenges and evaluates three graph neural network architectures for multi-omics (MO) integration based on graph-convolutional networks (GCN), graph-attention networks (GAT), and graph-transformer networks (GTN) for classifying 31 cancer types and normal tissues. To address the high-dimensionality of multi-omics data, we employed LASSO (Least Absolute Shrinkage and Selection Operator) regression for feature selection, leading to the creation of LASSO-MOGCN, LASSO-MOGAT, and LASSO-MOTGN models. Graph structures for the networks were constructed using gene correlation matrices and protein-protein interaction networks for multi-omics integration of messenger-RNA, micro-RNA, and DNA methylation data. Such data integration enables the networks to dynamically focus on important relationships between biological entities, improving both model performance and interpretability. Among the models, LASSO-MOGAT with a correlation-based graph structure achieved state-of-the-art accuracy (95.9%) and outperformed the LASSO-MOGCN and LASSO-MOTGN models in terms of precision, recall, and F1-score. Our findings demonstrate that integrating multi-omics data in graph-based architectures enhances cancer classification performance by uncovering distinct molecular patterns that contribute to a better understanding of cancer biology and potential biomarkers for disease progression.
LASSO-MOGAT: A Multi-Omics Graph Attention Framework for Cancer Classification
Aug 30, 2024



Abstract:The application of machine learning methods to analyze changes in gene expression patterns has recently emerged as a powerful approach in cancer research, enhancing our understanding of the molecular mechanisms underpinning cancer development and progression. Combining gene expression data with other types of omics data has been reported by numerous works to improve cancer classification outcomes. Despite these advances, effectively integrating high-dimensional multi-omics data and capturing the complex relationships across different biological layers remains challenging. This paper introduces LASSO-MOGAT (LASSO-Multi-Omics Gated ATtention), a novel graph-based deep learning framework that integrates messenger RNA, microRNA, and DNA methylation data to classify 31 cancer types. Utilizing differential expression analysis with LIMMA and LASSO regression for feature selection, and leveraging Graph Attention Networks (GATs) to incorporate protein-protein interaction (PPI) networks, LASSO-MOGAT effectively captures intricate relationships within multi-omics data. Experimental validation using five-fold cross-validation demonstrates the method's precision, reliability, and capacity for providing comprehensive insights into cancer molecular mechanisms. The computation of attention coefficients for the edges in the graph by the proposed graph-attention architecture based on protein-protein interactions proved beneficial for identifying synergies in multi-omics data for cancer classification.
A2DMN: Anatomy-Aware Dilated Multiscale Network for Breast Ultrasound Semantic Segmentation
Mar 22, 2024Abstract:In recent years, convolutional neural networks for semantic segmentation of breast ultrasound (BUS) images have shown great success; however, two major challenges still exist. 1) Most current approaches inherently lack the ability to utilize tissue anatomy, resulting in misclassified image regions. 2) They struggle to produce accurate boundaries due to the repeated down-sampling operations. To address these issues, we propose a novel breast anatomy-aware network for capturing fine image details and a new smoothness term that encodes breast anatomy. It incorporates context information across multiple spatial scales to generate more accurate semantic boundaries. Extensive experiments are conducted to compare the proposed method and eight state-of-the-art approaches using a BUS dataset with 325 images. The results demonstrate the proposed method significantly improves the segmentation of the muscle, mammary, and tumor classes and produces more accurate fine details of tissue boundaries.
Uncertainty Quantification in Multivariable Regression for Material Property Prediction with Bayesian Neural Networks
Nov 04, 2023Abstract:With the increased use of data-driven approaches and machine learning-based methods in material science, the importance of reliable uncertainty quantification (UQ) of the predicted variables for informed decision-making cannot be overstated. UQ in material property prediction poses unique challenges, including the multi-scale and multi-physics nature of advanced materials, intricate interactions between numerous factors, limited availability of large curated datasets for model training, etc. Recently, Bayesian Neural Networks (BNNs) have emerged as a promising approach for UQ, offering a probabilistic framework for capturing uncertainties within neural networks. In this work, we introduce an approach for UQ within physics-informed BNNs, which integrates knowledge from governing laws in material modeling to guide the models toward physically consistent predictions. To evaluate the effectiveness of this approach, we present case studies for predicting the creep rupture life of steel alloys. Experimental validation with three datasets of collected measurements from creep tests demonstrates the ability of BNNs to produce accurate point and uncertainty estimates that are competitive or exceed the performance of the conventional method of Gaussian Process Regression. Similarly, we evaluated the suitability of BNNs for UQ in an active learning application and reported competitive performance. The most promising framework for creep life prediction is BNNs based on Markov Chain Monte Carlo approximation of the posterior distribution of network parameters, as it provided more reliable results in comparison to BNNs based on variational inference approximation or related NNs with probabilistic outputs. The codes are available at: https://github.com/avakanski/Creep-uncertainty-quantification.
Review of Machine Learning Methods for Additive Manufacturing of Functionally Graded Materials
Sep 28, 2023Abstract:Additive manufacturing has revolutionized the manufacturing of complex parts by enabling direct material joining and offers several advantages such as cost-effective manufacturing of complex parts, reducing manufacturing waste, and opening new possibilities for manufacturing automation. One group of materials for which additive manufacturing holds great potential for enhancing component performance and properties is Functionally Graded Materials (FGMs). FGMs are advanced composite materials that exhibit smoothly varying properties making them desirable for applications in aerospace, automobile, biomedical, and defense industries. Such composition differs from traditional composite materials, since the location-dependent composition changes gradually in FGMs, leading to enhanced properties. Recently, machine learning techniques have emerged as a promising means for fabrication of FGMs through optimizing processing parameters, improving product quality, and detecting manufacturing defects. This paper first provides a brief literature review of works related to FGM fabrication, followed by reviewing works on employing machine learning in additive manufacturing, Afterward, we provide an overview of published works in the literature related to the application of machine learning methods in Directed Energy Deposition and for fabrication of FGMs.
Post-Hoc Explainability of BI-RADS Descriptors in a Multi-task Framework for Breast Cancer Detection and Segmentation
Aug 27, 2023



Abstract:Despite recent medical advancements, breast cancer remains one of the most prevalent and deadly diseases among women. Although machine learning-based Computer-Aided Diagnosis (CAD) systems have shown potential to assist radiologists in analyzing medical images, the opaque nature of the best-performing CAD systems has raised concerns about their trustworthiness and interpretability. This paper proposes MT-BI-RADS, a novel explainable deep learning approach for tumor detection in Breast Ultrasound (BUS) images. The approach offers three levels of explanations to enable radiologists to comprehend the decision-making process in predicting tumor malignancy. Firstly, the proposed model outputs the BI-RADS categories used for BUS image analysis by radiologists. Secondly, the model employs multi-task learning to concurrently segment regions in images that correspond to tumors. Thirdly, the proposed approach outputs quantified contributions of each BI-RADS descriptor toward predicting the benign or malignant class using post-hoc explanations with Shapley Values.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge