Brian McClannahan
Classification of Long Noncoding RNA Elements Using Deep Convolutional Neural Networks and Siamese Networks
Feb 10, 2021



Abstract:In the last decade, the discovery of noncoding RNA(ncRNA) has exploded. Classifying these ncRNA is critical todetermining their function. This thesis proposes a new methodemploying deep convolutional neural networks (CNNs) to classifyncRNA sequences. To this end, this paper first proposes anefficient approach to convert the RNA sequences into imagescharacterizing their base-pairing probability. As a result, clas-sifying RNA sequences is converted to an image classificationproblem that can be efficiently solved by available CNN-basedclassification models. This research also considers the foldingpotential of the ncRNAs in addition to their primary sequence.Based on the proposed approach, a benchmark image classifi-cation dataset is generated from the RFAM database of ncRNAsequences. In addition, three classical CNN models and threeSiamese network models have been implemented and comparedto demonstrate the superior performance and efficiency of theproposed approach. Extensive experimental results show thegreat potential of using deep learning approaches for RNAclassification.
Classification of Noncoding RNA Elements Using Deep Convolutional Neural Networks
Aug 24, 2020

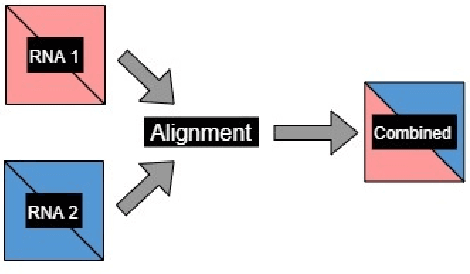
Abstract:The paper proposes to employ deep convolutional neural networks (CNNs) to classify noncoding RNA (ncRNA) sequences. To this end, we first propose an efficient approach to convert the RNA sequences into images characterizing their base-pairing probability. As a result, classifying RNA sequences is converted to an image classification problem that can be efficiently solved by available CNN-based classification models. The paper also considers the folding potential of the ncRNAs in addition to their primary sequence. Based on the proposed approach, a benchmark image classification dataset is generated from the RFAM database of ncRNA sequences. In addition, three classical CNN models have been implemented and compared to demonstrate the superior performance and efficiency of the proposed approach. Extensive experimental results show the great potential of using deep learning approaches for RNA classification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge