Boyu Ren
Multi-source domain adaptation for regression
Dec 09, 2023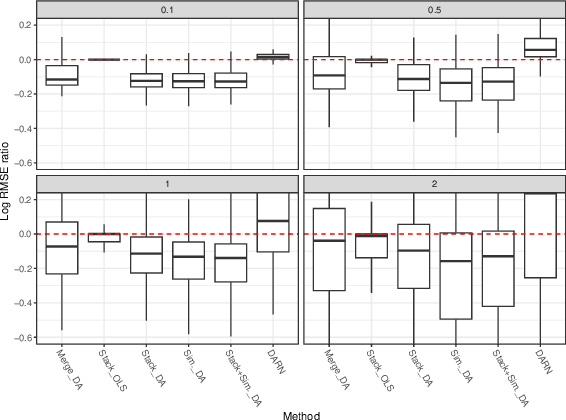
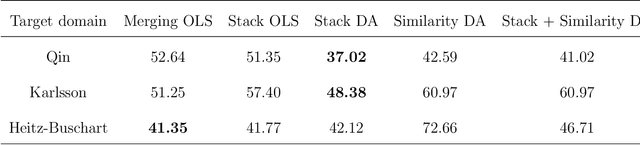
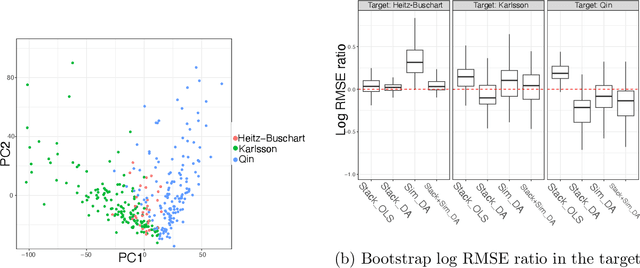
Abstract:Multi-source domain adaptation (DA) aims at leveraging information from more than one source domain to make predictions in a target domain, where different domains may have different data distributions. Most existing methods for multi-source DA focus on classification problems while there is only limited investigation in the regression settings. In this paper, we fill in this gap through a two-step procedure. First, we extend a flexible single-source DA algorithm for classification through outcome-coarsening to enable its application to regression problems. We then augment our single-source DA algorithm for regression with ensemble learning to achieve multi-source DA. We consider three learning paradigms in the ensemble algorithm, which combines linearly the target-adapted learners trained with each source domain: (i) a multi-source stacking algorithm to obtain the ensemble weights; (ii) a similarity-based weighting where the weights reflect the quality of DA of each target-adapted learner; and (iii) a combination of the stacking and similarity weights. We illustrate the performance of our algorithms with simulations and a data application where the goal is to predict High-density lipoprotein (HDL) cholesterol levels using gut microbiome. We observe a consistent improvement in prediction performance of our multi-source DA algorithm over the routinely used methods in all these scenarios.
Multi-study R-learner for Heterogeneous Treatment Effect Estimation
Jun 16, 2023Abstract:Estimating heterogeneous treatment effects is crucial for informing personalized treatment strategies and policies. While multiple studies can improve the accuracy and generalizability of results, leveraging them for estimation is statistically challenging. Existing approaches often assume identical heterogeneous treatment effects across studies, but this may be violated due to various sources of between-study heterogeneity, including differences in study design, confounders, and sample characteristics. To this end, we propose a unifying framework for multi-study heterogeneous treatment effect estimation that is robust to between-study heterogeneity in the nuisance functions and treatment effects. Our approach, the multi-study R-learner, extends the R-learner to obtain principled statistical estimation with modern machine learning (ML) in the multi-study setting. The multi-study R-learner is easy to implement and flexible in its ability to incorporate ML for estimating heterogeneous treatment effects, nuisance functions, and membership probabilities, which borrow strength across heterogeneous studies. It achieves robustness in confounding adjustment through its loss function and can leverage both randomized controlled trials and observational studies. We provide asymptotic guarantees for the proposed method in the case of series estimation and illustrate using real cancer data that it has the lowest estimation error compared to existing approaches in the presence of between-study heterogeneity.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge