Boudewijn Lelieveldt
Incorporating Texture Information into Dimensionality Reduction for High-Dimensional Images
Mar 02, 2022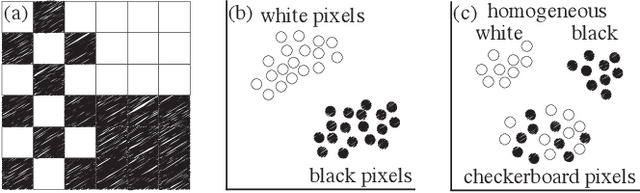
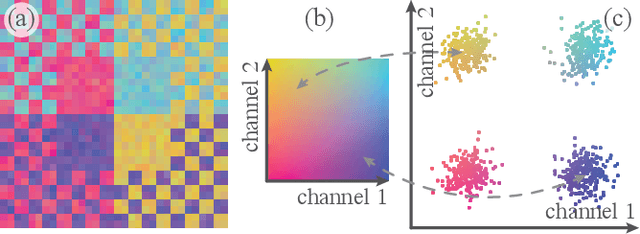
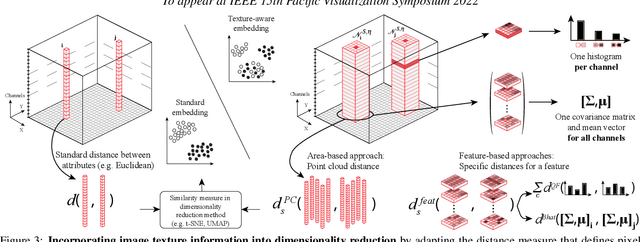
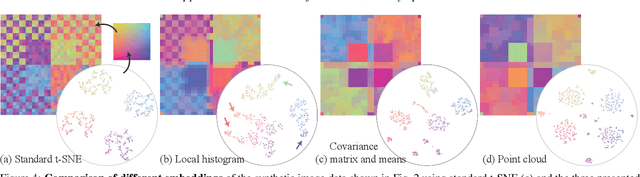
Abstract:High-dimensional imaging is becoming increasingly relevant in many fields from astronomy and cultural heritage to systems biology. Visual exploration of such high-dimensional data is commonly facilitated by dimensionality reduction. However, common dimensionality reduction methods do not include spatial information present in images, such as local texture features, into the construction of low-dimensional embeddings. Consequently, exploration of such data is typically split into a step focusing on the attribute space followed by a step focusing on spatial information, or vice versa. In this paper, we present a method for incorporating spatial neighborhood information into distance-based dimensionality reduction methods, such as t-Distributed Stochastic Neighbor Embedding (t-SNE). We achieve this by modifying the distance measure between high-dimensional attribute vectors associated with each pixel such that it takes the pixel's spatial neighborhood into account. Based on a classification of different methods for comparing image patches, we explore a number of different approaches. We compare these approaches from a theoretical and experimental point of view. Finally, we illustrate the value of the proposed methods by qualitative and quantitative evaluation on synthetic data and two real-world use cases.
Hierarchical stochastic neighbor embedding as a tool for visualizing the encoding capability of magnetic resonance fingerprinting dictionaries
Oct 07, 2019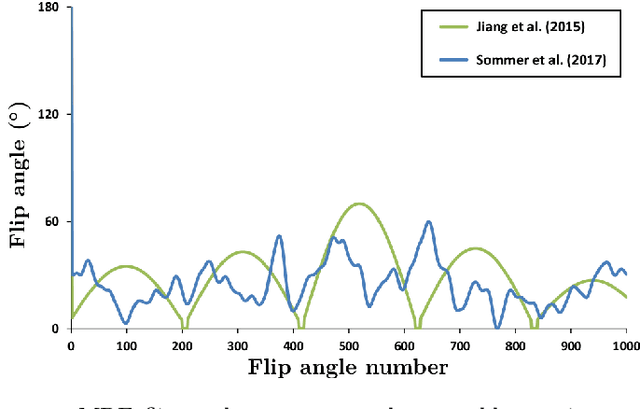
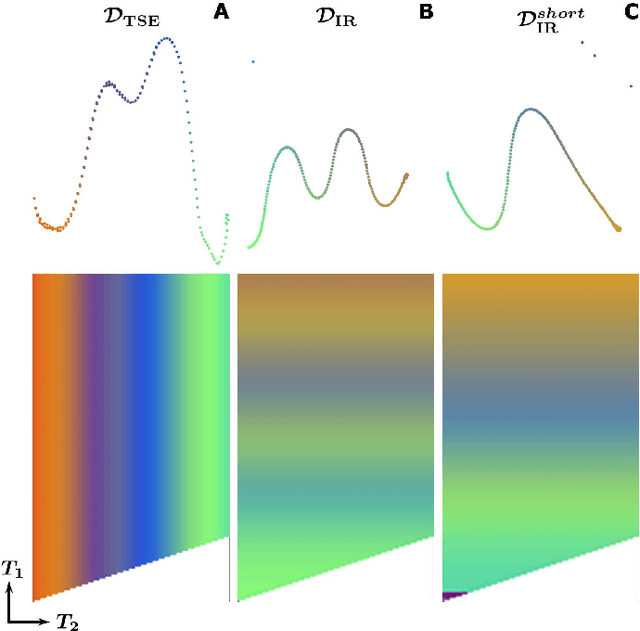
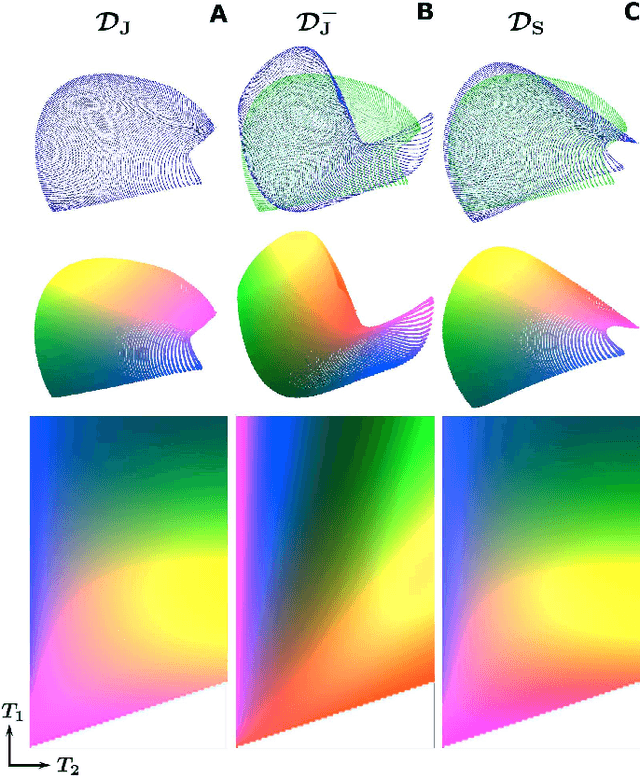
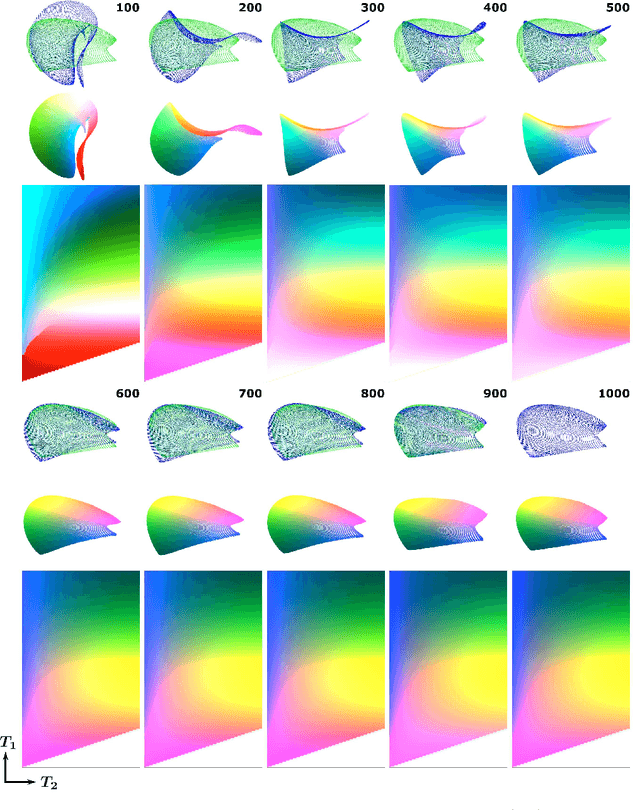
Abstract:In Magnetic Resonance Fingerprinting (MRF) the quality of the estimated parameter maps depends on the encoding capability of the variable flip angle train. In this work we show how the dimensionality reduction technique Hierarchical Stochastic Neighbor Embedding (HSNE) can be used to obtain insight into the encoding capability of different MRF sequences. Embedding high-dimensional MRF dictionaries into a lower-dimensional space and visualizing them with colors, being a surrogate for location in low-dimensional space, provides a comprehensive overview of particular dictionaries and, in addition, enables comparison of different sequences. Dictionaries for various sequences and sequence lengths were compared to each other, and the effect of transmit field variations on the encoding capability was assessed. Clear differences in encoding capability were observed between different sequences, and HSNE results accurately reflect those obtained from an MRF matching simulation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge