Biao Jie
Self-attention based high order sequence feature reconstruction of dynamic functional connectivity networks with rs-fMRI for brain disease classification
Nov 19, 2022Abstract:Dynamic functional connectivity networks (dFCN) based on rs-fMRI have demonstrated tremendous potential for brain function analysis and brain disease classification. Recently, studies have applied deep learning techniques (i.e., convolutional neural network, CNN) to dFCN classification, and achieved better performance than the traditional machine learning methods. Nevertheless, previous deep learning methods usually perform successive convolutional operations on the input dFCNs to obtain high-order brain network aggregation features, extracting them from each sliding window using a series split, which may neglect non-linear correlations among different regions and the sequentiality of information. Thus, important high-order sequence information of dFCNs, which could further improve the classification performance, is ignored in these studies. Nowadays, inspired by the great success of Transformer in natural language processing and computer vision, some latest work has also emerged on the application of Transformer for brain disease diagnosis based on rs-fMRI data. Although Transformer is capable of capturing non-linear correlations, it lacks accounting for capturing local spatial feature patterns and modelling the temporal dimension due to parallel computing, even equipped with a positional encoding technique. To address these issues, we propose a self-attention (SA) based convolutional recurrent network (SA-CRN) learning framework for brain disease classification with rs-fMRI data. The experimental results on a public dataset (i.e., ADNI) demonstrate the effectiveness of our proposed SA-CRN method.
Ordinal Pattern Kernel for Brain Connectivity Network Classification
Aug 18, 2020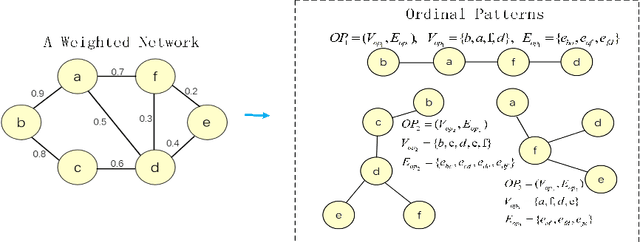
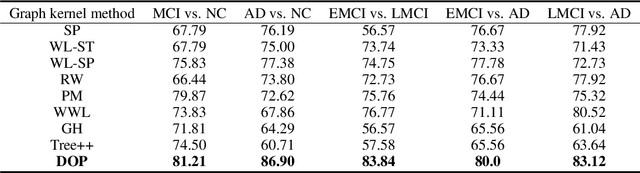
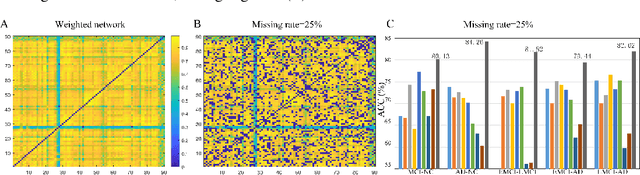
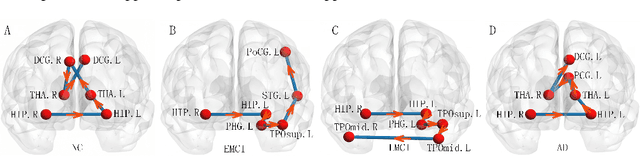
Abstract:Brain connectivity networks, which characterize the functional or structural interaction of brain regions, has been widely used for brain disease classification. Kernel-based method, such as graph kernel (i.e., kernel defined on graphs), has been proposed for measuring the similarity of brain networks, and yields the promising classification performance. However, most of graph kernels are built on unweighted graph (i.e., network) with edge present or not, and neglecting the valuable weight information of edges in brain connectivity network, with edge weights conveying the strengths of temporal correlation or fiber connection between brain regions. Accordingly, in this paper, we present an ordinal pattern kernel for brain connectivity network classification. Different with existing graph kernels that measures the topological similarity of unweighted graphs, the proposed ordinal pattern kernels calculate the similarity of weighted networks by comparing ordinal patterns from weighted networks. To evaluate the effectiveness of the proposed ordinal kernel, we further develop a depth-first-based ordinal pattern kernel, and perform extensive experiments in a real dataset of brain disease from ADNI database. The results demonstrate that our proposed ordinal pattern kernel can achieve better classification performance compared with state-of-the-art graph kernels.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge