Bharath Srinivas Prabakaran
Exploring Weakly Supervised Semantic Segmentation Ensembles for Medical Imaging Systems
Mar 16, 2023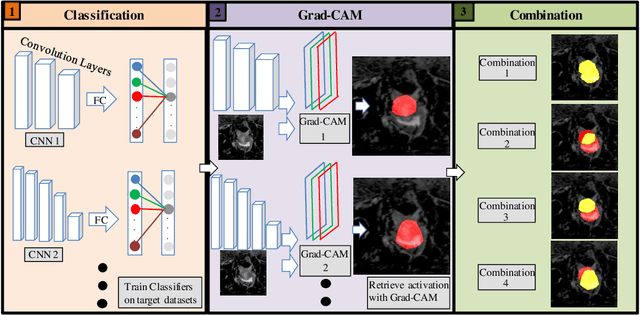
Abstract:Reliable classification and detection of certain medical conditions, in images, with state-of-the-art semantic segmentation networks, require vast amounts of pixel-wise annotation. However, the public availability of such datasets is minimal. Therefore, semantic segmentation with image-level labels presents a promising alternative to this problem. Nevertheless, very few works have focused on evaluating this technique and its applicability to the medical sector. Due to their complexity and the small number of training examples in medical datasets, classifier-based weakly supervised networks like class activation maps (CAMs) struggle to extract useful information from them. However, most state-of-the-art approaches rely on them to achieve their improvements. Therefore, we propose a framework that can still utilize the low-quality CAM predictions of complicated datasets to improve the accuracy of our results. Our framework achieves that by first utilizing lower threshold CAMs to cover the target object with high certainty; second, by combining multiple low-threshold CAMs that even out their errors while highlighting the target object. We performed exhaustive experiments on the popular multi-modal BRATS and prostate DECATHLON segmentation challenge datasets. Using the proposed framework, we have demonstrated an improved dice score of up to 8% on BRATS and 6% on DECATHLON datasets compared to the previous state-of-the-art.
Image Label based Semantic Segmentation Framework using Object Perimeters
Mar 14, 2023Abstract:Achieving high-quality semantic segmentation predictions using only image-level labels enables a new level of real-world applicability. Although state-of-the-art networks deliver reliable predictions, the amount of handcrafted pixel-wise annotations to enable these results are not feasible in many real-world applications. Hence, several works have already targeted this bottleneck, using classifier-based networks like Class Activation Maps (CAMs) as a base. Addressing CAM's weaknesses of fuzzy borders and incomplete predictions, state-of-the-art approaches rely only on adding regulations to the classifier loss or using pixel-similarity-based refinement after the fact. We propose a framework that introduces an additional module using object perimeters for improved saliency. We define object perimeter information as the line separating the object and background. Our new PerimeterFit module will be applied to pre-refine the CAM predictions before using the pixel-similarity-based network. In this way, our PerimeterFit increases the quality of the CAM prediction while simultaneously improving the false negative rate. We investigated a wide range of state-of-the-art unsupervised semantic segmentation networks and edge detection techniques to create useful perimeter maps, which enable our framework to predict object locations with sharper perimeters. We achieved up to 1.5\% improvement over frameworks without our PerimeterFit module. We conduct an exhaustive analysis to illustrate that our framework enhances existing state-of-the-art frameworks for image-level-based semantic segmentation. The framework is open-source and accessible online at https://github.com/ErikOstrowski/Perimeter-based-Semantic-Segmentation.
FPUS23: An Ultrasound Fetus Phantom Dataset with Deep Neural Network Evaluations for Fetus Orientations, Fetal Planes, and Anatomical Features
Mar 14, 2023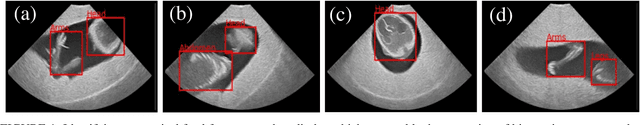
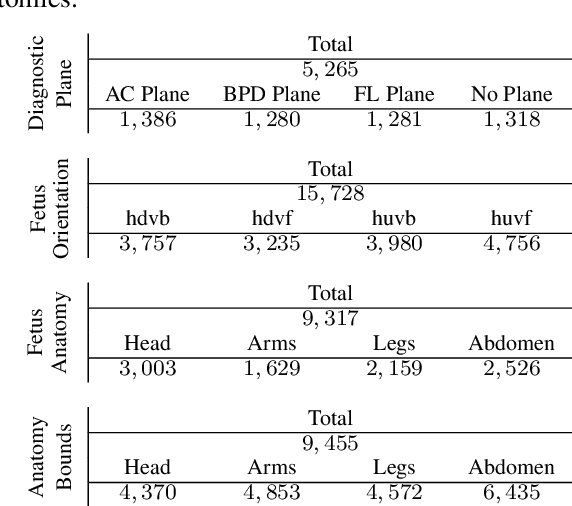
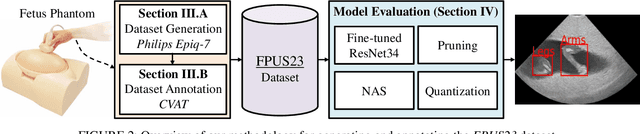
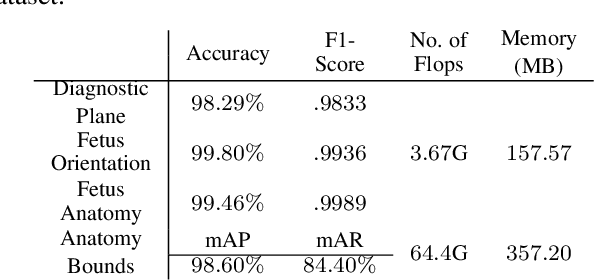
Abstract:Ultrasound imaging is one of the most prominent technologies to evaluate the growth, progression, and overall health of a fetus during its gestation. However, the interpretation of the data obtained from such studies is best left to expert physicians and technicians who are trained and well-versed in analyzing such images. To improve the clinical workflow and potentially develop an at-home ultrasound-based fetal monitoring platform, we present a novel fetus phantom ultrasound dataset, FPUS23, which can be used to identify (1) the correct diagnostic planes for estimating fetal biometric values, (2) fetus orientation, (3) their anatomical features, and (4) bounding boxes of the fetus phantom anatomies at 23 weeks gestation. The entire dataset is composed of 15,728 images, which are used to train four different Deep Neural Network models, built upon a ResNet34 backbone, for detecting aforementioned fetus features and use-cases. We have also evaluated the models trained using our FPUS23 dataset, to show that the information learned by these models can be used to substantially increase the accuracy on real-world ultrasound fetus datasets. We make the FPUS23 dataset and the pre-trained models publicly accessible at https://github.com/bharathprabakaran/FPUS23, which will further facilitate future research on fetal ultrasound imaging and analysis.
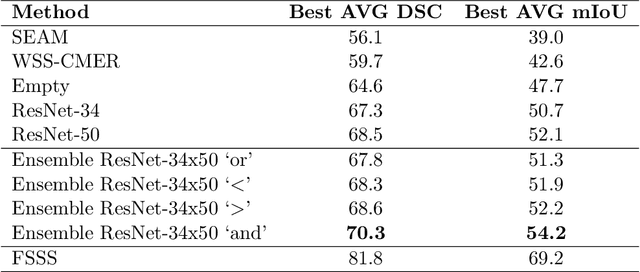
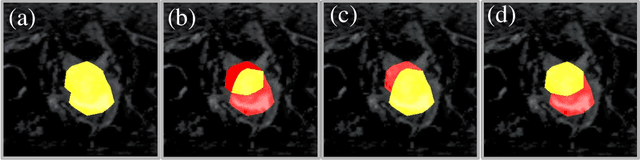
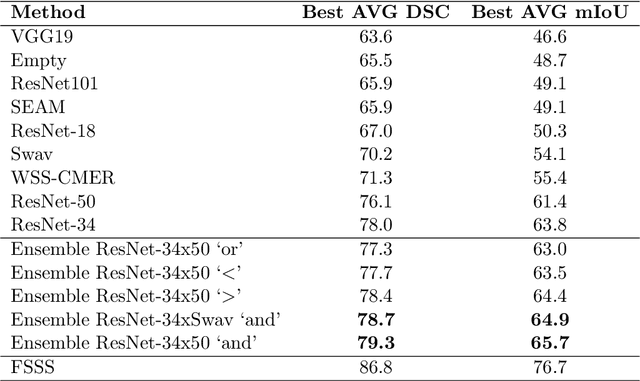
BoundaryCAM: A Boundary-based Refinement Framework for Weakly Supervised Semantic Segmentation of Medical Images
Mar 14, 2023



Abstract:Weakly Supervised Semantic Segmentation (WSSS) with only image-level supervision is a promising approach to deal with the need for Segmentation networks, especially for generating a large number of pixel-wise masks in a given dataset. However, most state-of-the-art image-level WSSS techniques lack an understanding of the geometric features embedded in the images since the network cannot derive any object boundary information from just image-level labels. We define a boundary here as the line separating an object and its background, or two different objects. To address this drawback, we propose our novel BoundaryCAM framework, which deploys state-of-the-art class activation maps combined with various post-processing techniques in order to achieve fine-grained higher-accuracy segmentation masks. To achieve this, we investigate a state-of-the-art unsupervised semantic segmentation network that can be used to construct a boundary map, which enables BoundaryCAM to predict object locations with sharper boundaries. By applying our method to WSSS predictions, we were able to achieve up to 10% improvements even to the benefit of the current state-of-the-art WSSS methods for medical imaging. The framework is open-source and accessible online at https://github.com/bharathprabakaran/BoundaryCAM.
UnbiasedNets: A Dataset Diversification Framework for Robustness Bias Alleviation in Neural Networks
Mar 13, 2023Abstract:Performance of trained neural network (NN) models, in terms of testing accuracy, has improved remarkably over the past several years, especially with the advent of deep learning. However, even the most accurate NNs can be biased toward a specific output classification due to the inherent bias in the available training datasets, which may propagate to the real-world implementations. This paper deals with the robustness bias, i.e., the bias exhibited by the trained NN by having a significantly large robustness to noise for a certain output class, as compared to the remaining output classes. The bias is shown to result from imbalanced datasets, i.e., the datasets where all output classes are not equally represented. Towards this, we propose the UnbiasedNets framework, which leverages K-means clustering and the NN's noise tolerance to diversify the given training dataset, even from relatively smaller datasets. This generates balanced datasets and reduces the bias within the datasets themselves. To the best of our knowledge, this is the first framework catering to the robustness bias problem in NNs. We use real-world datasets to demonstrate the efficacy of the UnbiasedNets for data diversification, in case of both binary and multi-label classifiers. The results are compared to well-known tools aimed at generating balanced datasets, and illustrate how existing works have limited success while addressing the robustness bias. In contrast, UnbiasedNets provides a notable improvement over existing works, while even reducing the robustness bias significantly in some cases, as observed by comparing the NNs trained on the diversified and original datasets.
* Springer Machine Learning 2023
BioNetExplorer: Architecture-Space Exploration of Bio-Signal Processing Deep Neural Networks for Wearables
Sep 07, 2021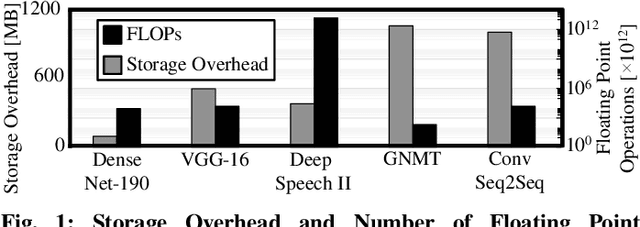
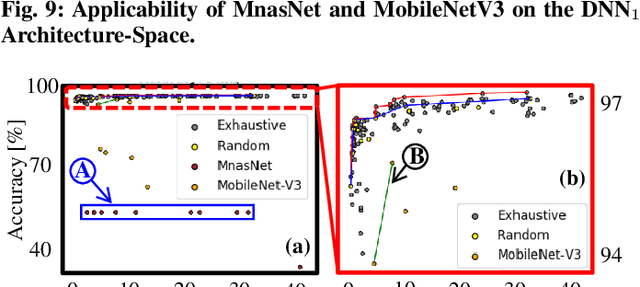
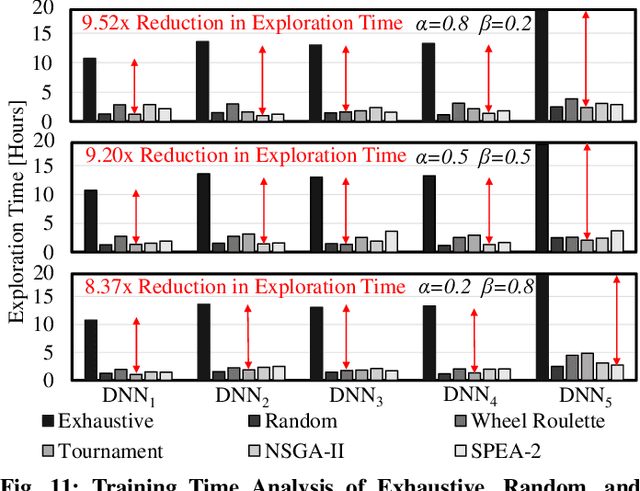
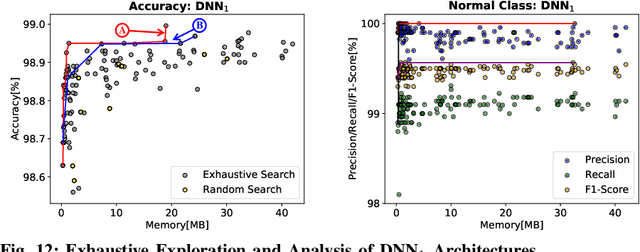
Abstract:In this work, we propose the BioNetExplorer framework to systematically generate and explore multiple DNN architectures for bio-signal processing in wearables. Our framework adapts key neural architecture parameters to search for an embedded DNN with a low hardware overhead, which can be deployed in wearable edge devices to analyse the bio-signal data and to extract the relevant information, such as arrhythmia and seizure. Our framework also enables hardware-aware DNN architecture search using genetic algorithms by imposing user requirements and hardware constraints (storage, FLOPs, etc.) during the exploration stage, thereby limiting the number of networks explored. Moreover, BioNetExplorer can also be used to search for DNNs based on the user-required output classes; for instance, a user might require a specific output class due to genetic predisposition or a pre-existing heart condition. The use of genetic algorithms reduces the exploration time, on average, by 9x, compared to exhaustive exploration. We are successful in identifying Pareto-optimal designs, which can reduce the storage overhead of the DNN by ~30MB for a quality loss of less than 0.5%. To enable low-cost embedded DNNs, BioNetExplorer also employs different model compression techniques to further reduce the storage overhead of the network by up to 53x for a quality loss of <0.2%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge