Erik Ostrowski
Embedded Deployment of Semantic Segmentation in Medicine through Low-Resolution Inputs
Mar 08, 2024



Abstract:When deploying neural networks in real-life situations, the size and computational effort are often the limiting factors. This is especially true in environments where big, expensive hardware is not affordable, like in embedded medical devices, where budgets are often tight. State-of-the-art proposed multiple different lightweight solutions for such use cases, mostly by changing the base model architecture, not taking the input and output resolution into consideration. In this paper, we propose our architecture that takes advantage of the fact that in hardware-limited environments, we often refrain from using the highest available input resolutions to guarantee a higher throughput. Although using lower-resolution input leads to a significant reduction in computing and memory requirements, it may also incur reduced prediction quality. Our architecture addresses this problem by exploiting the fact that we can still utilize high-resolution ground-truths in training. The proposed model inputs lower-resolution images and high-resolution ground truths, which can improve the prediction quality by 5.5% while adding less than 200 parameters to the model. %reducing the frames per second only from 25 to 20. We conduct an extensive analysis to illustrate that our architecture enhances existing state-of-the-art frameworks for lightweight semantic segmentation of cancer in MRI images. We also tested the deployment speed of state-of-the-art lightweight networks and our architecture on Nvidia's Jetson Nano to emulate deployment in resource-constrained embedded scenarios.
Exploring Weakly Supervised Semantic Segmentation Ensembles for Medical Imaging Systems
Mar 16, 2023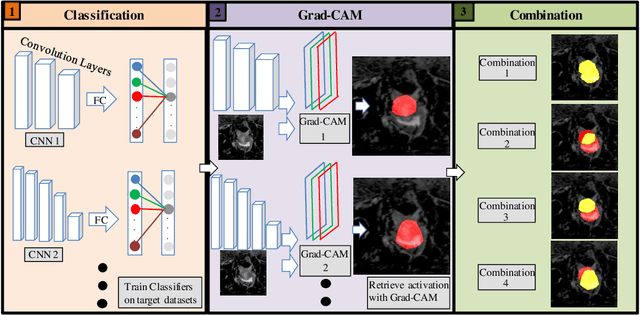
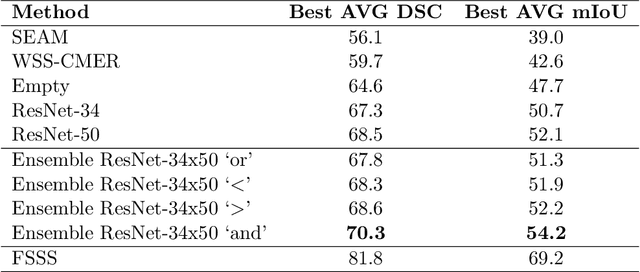
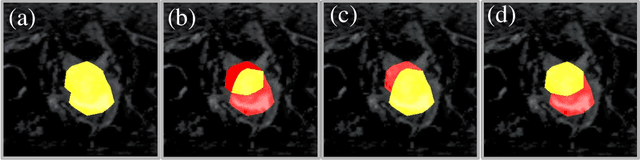
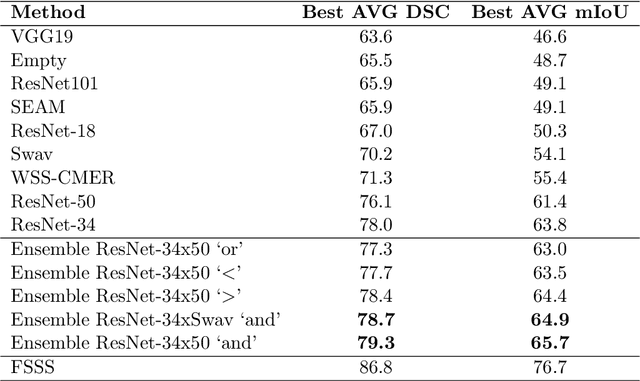
Abstract:Reliable classification and detection of certain medical conditions, in images, with state-of-the-art semantic segmentation networks, require vast amounts of pixel-wise annotation. However, the public availability of such datasets is minimal. Therefore, semantic segmentation with image-level labels presents a promising alternative to this problem. Nevertheless, very few works have focused on evaluating this technique and its applicability to the medical sector. Due to their complexity and the small number of training examples in medical datasets, classifier-based weakly supervised networks like class activation maps (CAMs) struggle to extract useful information from them. However, most state-of-the-art approaches rely on them to achieve their improvements. Therefore, we propose a framework that can still utilize the low-quality CAM predictions of complicated datasets to improve the accuracy of our results. Our framework achieves that by first utilizing lower threshold CAMs to cover the target object with high certainty; second, by combining multiple low-threshold CAMs that even out their errors while highlighting the target object. We performed exhaustive experiments on the popular multi-modal BRATS and prostate DECATHLON segmentation challenge datasets. Using the proposed framework, we have demonstrated an improved dice score of up to 8% on BRATS and 6% on DECATHLON datasets compared to the previous state-of-the-art.
AutoEnsemble: Automated Ensemble Search Framework for Semantic Segmentation Using Image Labels
Mar 15, 2023



Abstract:A key bottleneck of employing state-of-the-art semantic segmentation networks in the real world is the availability of training labels. Standard semantic segmentation networks require massive pixel-wise annotated labels to reach state-of-the-art prediction quality. Hence, several works focus on semantic segmentation networks trained with only image-level annotations. However, when scrutinizing the state-of-the-art results in more detail, we notice that although they are very close to each other on average prediction quality, different approaches perform better in different classes while providing low quality in others. To address this problem, we propose a novel framework, AutoEnsemble, which employs an ensemble of the "pseudo-labels" for a given set of different segmentation techniques on a class-wise level. Pseudo-labels are the pixel-wise predictions of the image-level semantic segmentation frameworks used to train the final segmentation model. Our pseudo-labels seamlessly combine the strong points of multiple segmentation techniques approaches to reach superior prediction quality. We reach up to 2.4% improvement over AutoEnsemble's components. An exhaustive analysis was performed to demonstrate AutoEnsemble's effectiveness over state-of-the-art frameworks for image-level semantic segmentation.
Image Label based Semantic Segmentation Framework using Object Perimeters
Mar 14, 2023Abstract:Achieving high-quality semantic segmentation predictions using only image-level labels enables a new level of real-world applicability. Although state-of-the-art networks deliver reliable predictions, the amount of handcrafted pixel-wise annotations to enable these results are not feasible in many real-world applications. Hence, several works have already targeted this bottleneck, using classifier-based networks like Class Activation Maps (CAMs) as a base. Addressing CAM's weaknesses of fuzzy borders and incomplete predictions, state-of-the-art approaches rely only on adding regulations to the classifier loss or using pixel-similarity-based refinement after the fact. We propose a framework that introduces an additional module using object perimeters for improved saliency. We define object perimeter information as the line separating the object and background. Our new PerimeterFit module will be applied to pre-refine the CAM predictions before using the pixel-similarity-based network. In this way, our PerimeterFit increases the quality of the CAM prediction while simultaneously improving the false negative rate. We investigated a wide range of state-of-the-art unsupervised semantic segmentation networks and edge detection techniques to create useful perimeter maps, which enable our framework to predict object locations with sharper perimeters. We achieved up to 1.5\% improvement over frameworks without our PerimeterFit module. We conduct an exhaustive analysis to illustrate that our framework enhances existing state-of-the-art frameworks for image-level-based semantic segmentation. The framework is open-source and accessible online at https://github.com/ErikOstrowski/Perimeter-based-Semantic-Segmentation.
FPUS23: An Ultrasound Fetus Phantom Dataset with Deep Neural Network Evaluations for Fetus Orientations, Fetal Planes, and Anatomical Features
Mar 14, 2023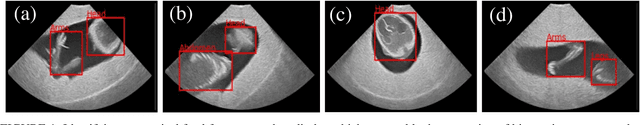
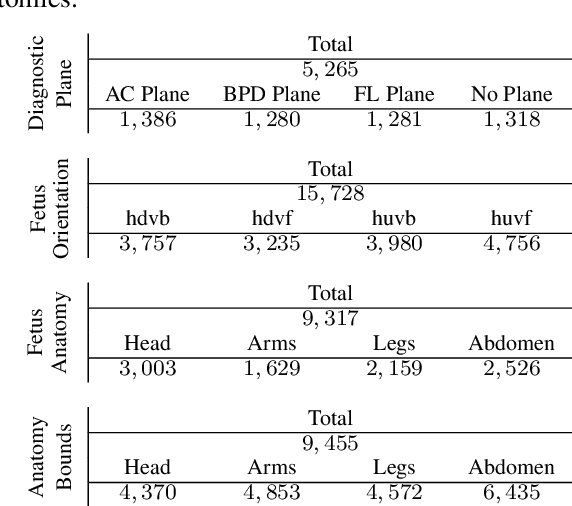
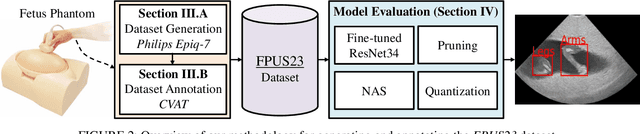
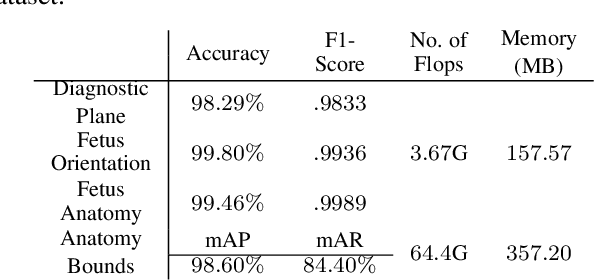
Abstract:Ultrasound imaging is one of the most prominent technologies to evaluate the growth, progression, and overall health of a fetus during its gestation. However, the interpretation of the data obtained from such studies is best left to expert physicians and technicians who are trained and well-versed in analyzing such images. To improve the clinical workflow and potentially develop an at-home ultrasound-based fetal monitoring platform, we present a novel fetus phantom ultrasound dataset, FPUS23, which can be used to identify (1) the correct diagnostic planes for estimating fetal biometric values, (2) fetus orientation, (3) their anatomical features, and (4) bounding boxes of the fetus phantom anatomies at 23 weeks gestation. The entire dataset is composed of 15,728 images, which are used to train four different Deep Neural Network models, built upon a ResNet34 backbone, for detecting aforementioned fetus features and use-cases. We have also evaluated the models trained using our FPUS23 dataset, to show that the information learned by these models can be used to substantially increase the accuracy on real-world ultrasound fetus datasets. We make the FPUS23 dataset and the pre-trained models publicly accessible at https://github.com/bharathprabakaran/FPUS23, which will further facilitate future research on fetal ultrasound imaging and analysis.
BoundaryCAM: A Boundary-based Refinement Framework for Weakly Supervised Semantic Segmentation of Medical Images
Mar 14, 2023



Abstract:Weakly Supervised Semantic Segmentation (WSSS) with only image-level supervision is a promising approach to deal with the need for Segmentation networks, especially for generating a large number of pixel-wise masks in a given dataset. However, most state-of-the-art image-level WSSS techniques lack an understanding of the geometric features embedded in the images since the network cannot derive any object boundary information from just image-level labels. We define a boundary here as the line separating an object and its background, or two different objects. To address this drawback, we propose our novel BoundaryCAM framework, which deploys state-of-the-art class activation maps combined with various post-processing techniques in order to achieve fine-grained higher-accuracy segmentation masks. To achieve this, we investigate a state-of-the-art unsupervised semantic segmentation network that can be used to construct a boundary map, which enables BoundaryCAM to predict object locations with sharper boundaries. By applying our method to WSSS predictions, we were able to achieve up to 10% improvements even to the benefit of the current state-of-the-art WSSS methods for medical imaging. The framework is open-source and accessible online at https://github.com/bharathprabakaran/BoundaryCAM.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge