Beth Ghavidel
Diffeomorphic Transformer-based Abdomen MRI-CT Deformable Image Registration
May 04, 2024


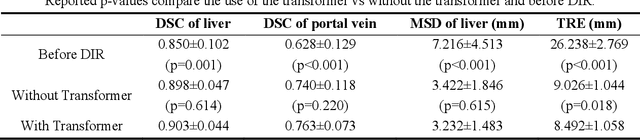
Abstract:This paper aims to create a deep learning framework that can estimate the deformation vector field (DVF) for directly registering abdominal MRI-CT images. The proposed method assumed a diffeomorphic deformation. By using topology-preserved deformation features extracted from the probabilistic diffeomorphic registration model, abdominal motion can be accurately obtained and utilized for DVF estimation. The model integrated Swin transformers, which have demonstrated superior performance in motion tracking, into the convolutional neural network (CNN) for deformation feature extraction. The model was optimized using a cross-modality image similarity loss and a surface matching loss. To compute the image loss, a modality-independent neighborhood descriptor (MIND) was used between the deformed MRI and CT images. The surface matching loss was determined by measuring the distance between the warped coordinates of the surfaces of contoured structures on the MRI and CT images. The deformed MRI image was assessed against the CT image using the target registration error (TRE), Dice similarity coefficient (DSC), and mean surface distance (MSD) between the deformed contours of the MRI image and manual contours of the CT image. When compared to only rigid registration, DIR with the proposed method resulted in an increase of the mean DSC values of the liver and portal vein from 0.850 and 0.628 to 0.903 and 0.763, a decrease of the mean MSD of the liver from 7.216 mm to 3.232 mm, and a decrease of the TRE from 26.238 mm to 8.492 mm. The proposed deformable image registration method based on a diffeomorphic transformer provides an effective and efficient way to generate an accurate DVF from an MRI-CT image pair of the abdomen. It could be utilized in the current treatment planning workflow for liver radiotherapy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge