Benjamin Lemasson
GIN
MARVEL: MR Fingerprinting with Additional micRoVascular Estimates using bidirectional LSTMs
Jul 15, 2024Abstract:The Magnetic Resonance Fingerprinting (MRF) approach aims to estimate multiple MR or physiological parameters simultaneously with a single fast acquisition sequence. Most of the MRF studies proposed so far have used simple MR sequence types to measure relaxation times (T1, T2). In that case, deep learning algorithms have been successfully used to speed up the reconstruction process. In theory, the MRF concept could be used with a variety of other MR sequence types and should be able to provide more information about the tissue microstructures. Yet, increasing the complexity of the numerical models often leads to prohibited simulation times, and estimating multiple parameters from one sequence implies new dictionary dimensions whose sizes become too large for standard computers and DL architectures.In this paper, we propose to analyze the MRF signal coming from a complex balance Steady-state free precession (bSSFP) type sequence to simultaneously estimate relaxometry maps (T1, T2), Field maps (B1, B0) as well as microvascular properties such as the local Cerebral Blood Volume (CBV) or the averaged vessel Radius (R).To bypass the curse of dimensionality, we propose an efficient way to simulate the MR signal coming from numerical voxels containing realistic microvascular networks as well as a Bidirectional Long Short-Term Memory network used for the matching process.On top of standard MRF maps, our results on 3 human volunteers suggest that our approach can quickly produce high-quality quantitative maps of microvascular parameters that are otherwise obtained using longer dedicated sequences and intravenous injection of a contrast agent. This approach could be used for the management of multiple pathologies and could be tuned to provide other types of microstructural information.
Enhancing MR vascular Fingerprinting through realistic microvascular geometries
May 26, 2023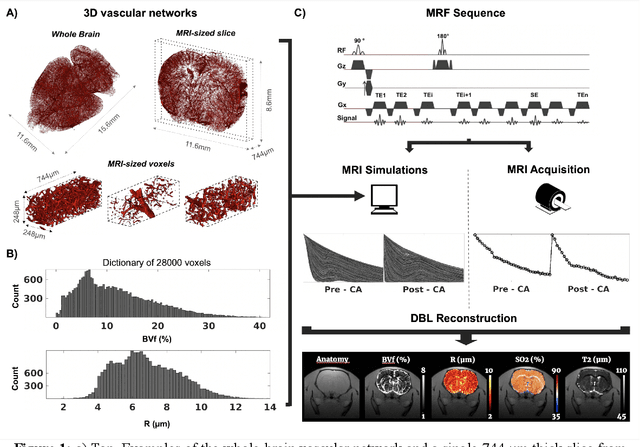



Abstract:MR vascular Fingerprinting proposes to use the MR Fingerprinting framework to quantitatively and simultaneously map several microvascular characteristics at a sub-voxel scale. The initial implementation assessed the local blood oxygenation saturation (SO 2), blood volume fraction (BVf) and vessel averaged radius (R) in humans and rodent brains using simple 2D representations of the vascular network during dictionary generation. In order to improve the results and possibly extend the approach to pathological environments and other biomarkers, we propose in this study to use 3D realistic vascular geometries in the numerical simulations. 28,000 different synthetic voxels containing vascular networks segmented from whole brain healthy mice microscopy images were created. A Bayesian-based regression model was used for map reconstruction. We show on 8 healthy and 9 tumor bearing rats that realistic vascular representations yield microvascular estimates in better agreement with the literature than 2D or 3D cylindrical models. Furthermore, tumoral blood oxygenation estimates obtained with the proposed approach are the only ones correlating with in vivo optic-fiber measurements performed in the same animals.
Neural parameters estimation for brain tumor growth modeling
Jul 01, 2019


Abstract:Understanding the dynamics of brain tumor progression is essential for optimal treatment planning. Cast in a mathematical formulation, it is typically viewed as evaluation of a system of partial differential equations, wherein the physiological processes that govern the growth of the tumor are considered. To personalize the model, i.e. find a relevant set of parameters, with respect to the tumor dynamics of a particular patient, the model is informed from empirical data, e.g., medical images obtained from diagnostic modalities, such as magnetic-resonance imaging. Existing model-observation coupling schemes require a large number of forward integrations of the biophysical model and rely on simplifying assumption on the functional form, linking the output of the model with the image information. In this work, we propose a learning-based technique for the estimation of tumor growth model parameters from medical scans. The technique allows for explicit evaluation of the posterior distribution of the parameters by sequentially training a mixture-density network, relaxing the constraint on the functional form and reducing the number of samples necessary to propagate through the forward model for the estimation. We test the method on synthetic and real scans of rats injected with brain tumors to calibrate the model and to predict tumor progression.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge