Babak Hossein Khalaj
Real-Time In-Cabin Driver Behavior Recognition on Low-Cost Edge Hardware
Dec 26, 2025Abstract:In-cabin Driver Monitoring Systems (DMS) must recognize distraction- and drowsiness-related behaviors with low latency under strict constraints on compute, power, and cost. We present a single-camera in-cabin driver behavior recognition system designed for deployment on two low-cost edge platforms: Raspberry Pi 5 (CPU-only) and Google Coral Edge TPU. The proposed pipeline combines (i) a compact per-frame vision model, (ii) a confounder-aware label design to reduce visually similar false positives, and (iii) a temporal decision head that triggers alerts only when predictions are both confident and sustained. The system covers 17 behavior classes, including multiple phone-use modes, eating/drinking, smoking, reaching behind, gaze/attention shifts, passenger interaction, grooming, control-panel interaction, yawning, and eyes-closed sleep. Training and evaluation use licensed datasets spanning diverse drivers, vehicles, and lighting conditions (details in Section 6), and we further validate runtime behavior in real in-vehicle tests. The optimized deployments achieve about 16 FPS on Raspberry Pi 5 with INT8 inference (per-frame latency under 60 ms) and about 25 FPS on Coral Edge TPU, enabling real-time monitoring and stable alert generation on inexpensive hardware. Finally, we discuss how reliable in-cabin human-state perception can serve as an upstream input for human-centered vehicle intelligence, including emerging agentic vehicle concepts.
Deep Joint Source Channel Coding for Secure End-to-End Image Transmission
Dec 22, 2024Abstract:Deep neural network (DNN)-based joint source and channel coding is proposed for end-to-end secure image transmission against multiple eavesdroppers. Both scenarios of colluding and non-colluding eavesdroppers are considered. Instead of idealistic assumptions of perfectly known and i.i.d. source and channel distributions, the proposed scheme assumes unknown source and channel statistics. The goal is to transmit images with minimum distortion, while simultaneously preventing eavesdroppers from inferring private attributes of images. Simultaneously generalizing the ideas of privacy funnel and wiretap coding, a multi-objective optimization framework is expressed that characterizes the trade-off between image reconstruction quality and information leakage to eavesdroppers, taking into account the structural similarity index (SSIM) for improving the perceptual quality of image reconstruction. Extensive experiments over CIFAR-10 and CelebFaces Attributes (CelebA) datasets, together with ablation studies are provided to highlight the performance gain in terms of SSIM, adversarial accuracy, and cross-entropy metric compared with benchmarks. Experiments show that the proposed scheme restrains the adversarially-trained eavesdroppers from intercepting privatized data for both cases of eavesdropping a common secret, as well as the case in which eavesdroppers are interested in different secrets. Furthermore, useful insights on the privacy-utility trade-off are also provided.
Secure Deep-JSCC Against Multiple Eavesdroppers
Aug 05, 2023Abstract:In this paper, a generalization of deep learning-aided joint source channel coding (Deep-JSCC) approach to secure communications is studied. We propose an end-to-end (E2E) learning-based approach for secure communication against multiple eavesdroppers over complex-valued fading channels. Both scenarios of colluding and non-colluding eavesdroppers are studied. For the colluding strategy, eavesdroppers share their logits to collaboratively infer private attributes based on ensemble learning method, while for the non-colluding setup they act alone. The goal is to prevent eavesdroppers from inferring private (sensitive) information about the transmitted images, while delivering the images to a legitimate receiver with minimum distortion. By generalizing the ideas of privacy funnel and wiretap channel coding, the trade-off between the image recovery at the legitimate node and the information leakage to the eavesdroppers is characterized. To solve this secrecy funnel framework, we implement deep neural networks (DNNs) to realize a data-driven secure communication scheme, without relying on a specific data distribution. Simulations over CIFAR-10 dataset verifies the secrecy-utility trade-off. Adversarial accuracy of eavesdroppers are also studied over Rayleigh fading, Nakagami-m, and AWGN channels to verify the generalization of the proposed scheme. Our experiments show that employing the proposed secure neural encoding can decrease the adversarial accuracy by 28%.
A Geometric Approach For Fully Automatic Chromosome Segmentation
Sep 01, 2014
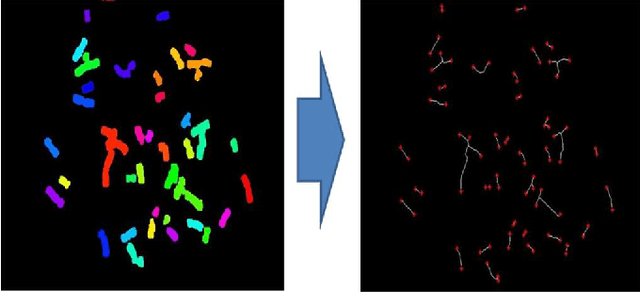

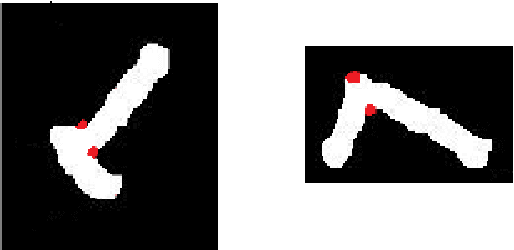
Abstract:A fundamental task in human chromosome analysis is chromosome segmentation. Segmentation plays an important role in chromosome karyotyping. The first step in segmentation is to remove intrusive objects such as stain debris and other noises. The next step is detection of touching and overlapping chromosomes, and the final step is separation of such chromosomes. Common methods for separation between touching chromosomes are interactive and require human intervention for correct separation between touching and overlapping chromosomes. In this paper, a geometric-based method is used for automatic detection of touching and overlapping chromosomes and separating them. The proposed scheme performs segmentation in two phases. In the first phase, chromosome clusters are detected using three geometric criteria, and in the second phase, chromosome clusters are separated using a cut-line. Most of earlier methods did not work properly in case of chromosome clusters that contained more than two chromosomes. Our method, on the other hand, is quite efficient in separation of such chromosome clusters. At each step, one separation will be performed and this algorithm is repeated until all individual chromosomes are separated. Another important point about the proposed method is that it uses the geometric features of chromosomes which are independent of the type of images and it can easily be applied to any type of images such as binary images and does not require multispectral images as well. We have applied our method to a database containing 62 touching and partially overlapping chromosomes and a success rate of 91.9% is achieved.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge