Austin J. Brockmeier
A Systematic Analysis of Out-of-Distribution Detection Under Representation and Training Paradigm Shifts
Nov 14, 2025Abstract:We present a systematic comparison of out-of-distribution (OOD) detection methods across CLIP-stratified regimes using AURC and AUGRC as primary metrics. Experiments cover two representation paradigms: CNNs trained from scratch and a fine-tuned Vision Transformer (ViT), evaluated on CIFAR-10/100, SuperCIFAR-100, and TinyImageNet. Using a multiple-comparison-controlled, rank-based pipeline (Friedman test with Conover-Holm post-hoc) and Bron-Kerbosch cliques, we find that the learned feature space largely determines OOD efficacy. For both CNNs and ViTs, probabilistic scores (e.g., MSR, GEN) dominate misclassification (ID) detection. Under stronger shifts, geometry-aware scores (e.g., NNGuide, fDBD, CTM) prevail on CNNs, whereas on ViTs GradNorm and KPCA Reconstruction Error remain consistently competitive. We further show a class-count-dependent trade-off for Monte-Carlo Dropout (MCD) and that a simple PCA projection improves several detectors. These results support a representation-centric view of OOD detection and provide statistically grounded guidance for method selection under distribution shift.
Measuring Chain-of-Thought Monitorability Through Faithfulness and Verbosity
Oct 31, 2025Abstract:Chain-of-thought (CoT) outputs let us read a model's step-by-step reasoning. Since any long, serial reasoning process must pass through this textual trace, the quality of the CoT is a direct window into what the model is thinking. This visibility could help us spot unsafe or misaligned behavior (monitorability), but only if the CoT is transparent about its internal reasoning (faithfulness). Fully measuring faithfulness is difficult, so researchers often focus on examining the CoT in cases where the model changes its answer after adding a cue to the input. This proxy finds some instances of unfaithfulness but loses information when the model maintains its answer, and does not investigate aspects of reasoning not tied to the cue. We extend these results to a more holistic sense of monitorability by introducing verbosity: whether the CoT lists every factor needed to solve the task. We combine faithfulness and verbosity into a single monitorability score that shows how well the CoT serves as the model's external `working memory', a property that many safety schemes based on CoT monitoring depend on. We evaluate instruction-tuned and reasoning models on BBH, GPQA, and MMLU. Our results show that models can appear faithful yet remain hard to monitor when they leave out key factors, and that monitorability differs sharply across model families. We release our evaluation code using the Inspect library to support reproducible future work.
Anomaly Detection via Autoencoder Composite Features and NCE
Feb 04, 2025



Abstract:Unsupervised anomaly detection is a challenging task. Autoencoders (AEs) or generative models are often employed to model the data distribution of normal inputs and subsequently identify anomalous, out-of-distribution inputs by high reconstruction error or low likelihood, respectively. However, AEs may generalize and achieve small reconstruction errors on abnormal inputs. We propose a decoupled training approach for anomaly detection that both an AE and a likelihood model trained with noise contrastive estimation (NCE). After training the AE, NCE estimates a probability density function, to serve as the anomaly score, on the joint space of the AE's latent representation combined with features of the reconstruction quality. To further reduce the false negative rate in NCE we systematically varying the reconstruction features to augment the training and optimize the contrastive Gaussian noise distribution. Experimental assessments on multiple benchmark datasets demonstrate that the proposed approach matches the performance of prevalent state-of-the-art anomaly detection algorithms.
Contrastive Learning to Fine-Tune Feature Extraction Models for the Visual Cortex
Oct 08, 2024Abstract:Predicting the neural response to natural images in the visual cortex requires extracting relevant features from the images and relating those feature to the observed responses. In this work, we optimize the feature extraction in order to maximize the information shared between the image features and the neural response across voxels in a given region of interest (ROI) extracted from the BOLD signal measured by fMRI. We adapt contrastive learning (CL) to fine-tune a convolutional neural network, which was pretrained for image classification, such that a mapping of a given image's features are more similar to the corresponding fMRI response than to the responses to other images. We exploit the recently released Natural Scenes Dataset (Allen et al., 2022) as organized for the Algonauts Project (Gifford et al., 2023), which contains the high-resolution fMRI responses of eight subjects to tens of thousands of naturalistic images. We show that CL fine-tuning creates feature extraction models that enable higher encoding accuracy in early visual ROIs as compared to both the pretrained network and a baseline approach that uses a regression loss at the output of the network to tune it for fMRI response encoding. We investigate inter-subject transfer of the CL fine-tuned models, including subjects from another, lower-resolution dataset (Gong et al., 2023). We also pool subjects for fine-tuning to further improve the encoding performance. Finally, we examine the performance of the fine-tuned models on common image classification tasks, explore the landscape of ROI-specific models by applying dimensionality reduction on the Bhattacharya dissimilarity matrix created using the predictions on those tasks (Mao et al., 2024), and investigate lateralization of the processing for early visual ROIs using salience maps of the classifiers built on the CL-tuned models.
DiME: Maximizing Mutual Information by a Difference of Matrix-Based Entropies
Jan 19, 2023Abstract:We introduce an information-theoretic quantity with similar properties to mutual information that can be estimated from data without making explicit assumptions on the underlying distribution. This quantity is based on a recently proposed matrix-based entropy that uses the eigenvalues of a normalized Gram matrix to compute an estimate of the eigenvalues of an uncentered covariance operator in a reproducing kernel Hilbert space. We show that a difference of matrix-based entropies (DiME) is well suited for problems involving maximization of mutual information between random variables. While many methods for such tasks can lead to trivial solutions, DiME naturally penalizes such outcomes. We provide several examples of use cases for the proposed quantity including a multi-view representation learning problem where DiME is used to encourage learning a shared representation among views with high mutual information. We also show the versatility of DiME by using it as objective function for a variety of tasks.
Exploring latent networks in resting-state fMRI using voxel-to-voxel causal modeling feature selection
Nov 15, 2021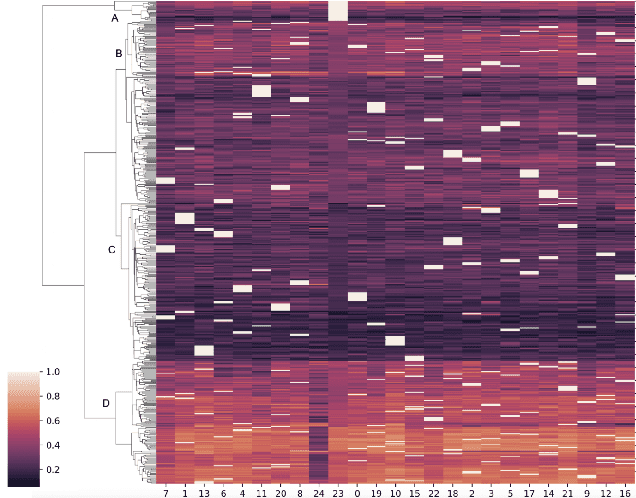
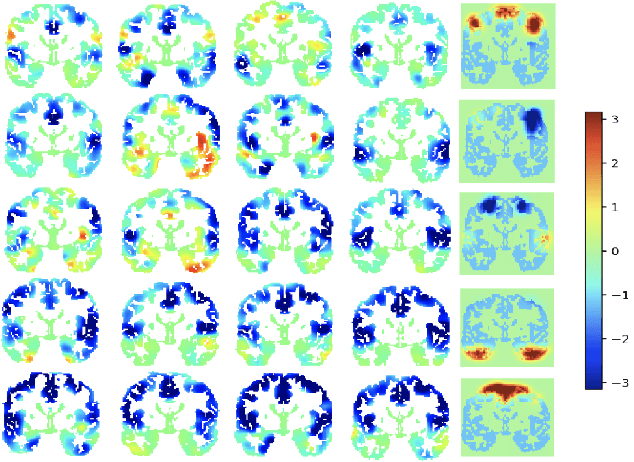
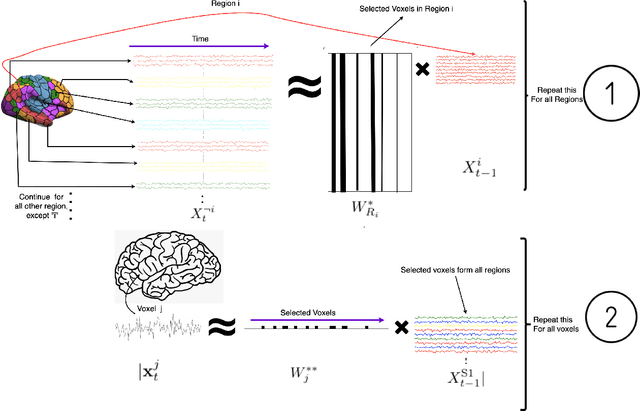
Abstract:Functional networks characterize the coordinated neural activity observed by functional neuroimaging. The prevalence of different networks during resting state periods provide useful features for predicting the trajectory of neurodegenerative diseases. Techniques for network estimation rely on statistical correlation or dependence between voxels. Due to the large number of voxels, rather than consider the voxel-to-voxel correlations between all voxels, a small set of seed voxels are chosen. Consequently, the network identification may depend on the selected seeds. As an alternative, we propose to fit first-order linear models with sparse priors on the coefficients to model activity across the entire set of cortical grey matter voxels as a linear combination of a smaller subset of voxels. We propose a two-stage algorithm for voxel subset selection that uses different sparsity-inducing regularization approaches to identify subject-specific causally predictive voxels. To reveal the functional networks among these voxels, we then apply independent component analysis (ICA) to model these voxels' signals as a mixture of latent sources each defining a functional network. Based on the inter-subject similarity of the sources' spatial patterns we identify independent sources that are well-matched across subjects but fail to match the independent sources from a group-based ICA. These are resting state networks, common across subjects that group ICA does not reveal. These complementary networks could help to better identify neurodegeneration, a task left for future work.
Shift-invariant waveform learning on epileptic ECoG
Aug 12, 2021


Abstract:Seizure detection algorithms must discriminate abnormal neuronal activity associated with a seizure from normal neural activity in a variety of conditions. Our approach is to seek spatiotemporal waveforms with distinct morphology in electrocorticographic (ECoG) recordings of epileptic patients that are indicative of a subsequent seizure (preictal) versus non-seizure segments (interictal). To find these waveforms we apply a shift-invariant k-means algorithm to segments of spatially filtered signals to learn codebooks of prototypical waveforms. The frequency of the cluster labels from the codebooks is then used to train a binary classifier that predicts the class (preictal or interictal) of a test ECoG segment. We use the Matthews correlation coefficient to evaluate the performance of the classifier and the quality of the codebooks. We found that our method finds recurrent non-sinusoidal waveforms that could be used to build interpretable features for seizure prediction and that are also physiologically meaningful.
Searching for waveforms on spatially-filtered epileptic ECoG
Mar 25, 2021



Abstract:Seizures are one of the defining symptoms in patients with epilepsy, and due to their unannounced occurrence, they can pose a severe risk for the individual that suffers it. New research efforts are showing a promising future for the prediction and preemption of imminent seizures, and with those efforts, a vast and diverse set of features have been proposed for seizure prediction algorithms. However, the data-driven discovery of nonsinusoidal waveforms for seizure prediction is lacking in the literature, which is in stark contrast with recent works that show the close connection between the waveform morphology of neural oscillations and the physiology and pathophysiology of the brain, and especially its use in effectively discriminating between normal and abnormal oscillations in electrocorticographic (ECoG) recordings of epileptic patients. Here, we explore a scalable, energy-guided waveform search strategy on spatially-projected continuous multi-day ECoG data sets. Our work shows that data-driven waveform learning methods have the potential to not only contribute features with predictive power for seizure prediction, but also to facilitate the discovery of oscillatory patterns that could contribute to our understanding of the pathophysiology and etiology of seizures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge