Hassan Baker
Exploring latent networks in resting-state fMRI using voxel-to-voxel causal modeling feature selection
Nov 15, 2021
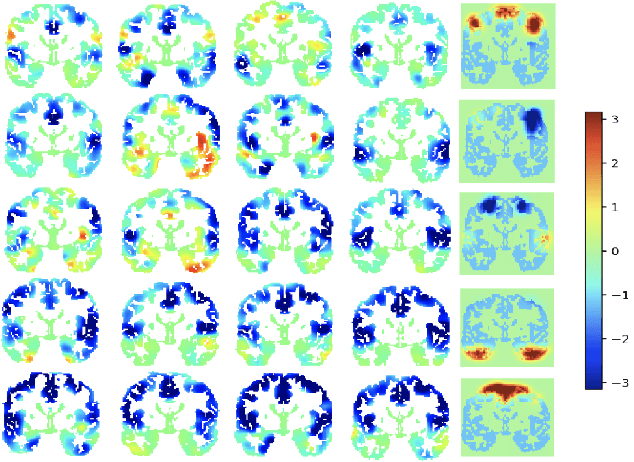
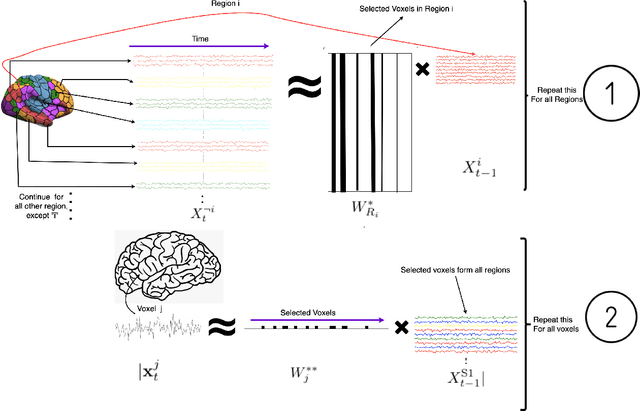
Abstract:Functional networks characterize the coordinated neural activity observed by functional neuroimaging. The prevalence of different networks during resting state periods provide useful features for predicting the trajectory of neurodegenerative diseases. Techniques for network estimation rely on statistical correlation or dependence between voxels. Due to the large number of voxels, rather than consider the voxel-to-voxel correlations between all voxels, a small set of seed voxels are chosen. Consequently, the network identification may depend on the selected seeds. As an alternative, we propose to fit first-order linear models with sparse priors on the coefficients to model activity across the entire set of cortical grey matter voxels as a linear combination of a smaller subset of voxels. We propose a two-stage algorithm for voxel subset selection that uses different sparsity-inducing regularization approaches to identify subject-specific causally predictive voxels. To reveal the functional networks among these voxels, we then apply independent component analysis (ICA) to model these voxels' signals as a mixture of latent sources each defining a functional network. Based on the inter-subject similarity of the sources' spatial patterns we identify independent sources that are well-matched across subjects but fail to match the independent sources from a group-based ICA. These are resting state networks, common across subjects that group ICA does not reveal. These complementary networks could help to better identify neurodegeneration, a task left for future work.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge