Asan Agibetov
Scalable and interpretable rule-based link prediction for large heterogeneous knowledge graphs
Dec 10, 2020



Abstract:Neural embedding-based machine learning models have shown promise for predicting novel links in biomedical knowledge graphs. Unfortunately, their practical utility is diminished by their lack of interpretability. Recently, the fully interpretable, rule-based algorithm AnyBURL yielded highly competitive results on many general-purpose link prediction benchmarks. However, its applicability to large-scale prediction tasks on complex biomedical knowledge bases is limited by long inference times and difficulties with aggregating predictions made by multiple rules. We improve upon AnyBURL by introducing the SAFRAN rule application framework which aggregates rules through a scalable clustering algorithm. SAFRAN yields new state-of-the-art results for fully interpretable link prediction on the established general-purpose benchmark FB15K-237 and the large-scale biomedical benchmark OpenBioLink. Furthermore, it exceeds the results of multiple established embedding-based algorithms on FB15K-237 and narrows the gap between rule-based and embedding-based algorithms on OpenBioLink. We also show that SAFRAN increases inference speeds by up to two orders of magnitude.
Graph embeddings via matrix factorization for link prediction: smoothing or truncating negatives?
Nov 16, 2020



Abstract:Link prediction -- the process of uncovering missing links in a complex network -- is an important problem in information sciences, with applications ranging from social sciences to molecular biology. Recent advances in neural graph embeddings have proposed an end-to-end way of learning latent vector representations of nodes, with successful application in link prediction tasks. Yet, our understanding of the internal mechanisms of such approaches has been rather limited, and only very recently we have witnessed the development of a very compelling connection to the mature matrix factorization theory. In this work, we make an important contribution to our understanding of the interplay between the skip-gram powered neural graph embedding algorithms and the matrix factorization via SVD. In particular, we show that the link prediction accuracy of graph embeddings strongly depends on the transformations of the original graph co-occurrence matrix that they decompose, sometimes resulting in staggering boosts of accuracy performance on link prediction tasks. Our improved approach to learning low-rank factorization embeddings that incorporate information from unlikely pairs of nodes yields results on par with the state-of-the-art link prediction performance achieved by a complex neural graph embedding model
Benchmarking neural embeddings for link prediction in knowledge graphs under semantic and structural changes
May 28, 2020



Abstract:Recently, link prediction algorithms based on neural embeddings have gained tremendous popularity in the Semantic Web community, and are extensively used for knowledge graph completion. While algorithmic advances have strongly focused on efficient ways of learning embeddings, fewer attention has been drawn to the different ways their performance and robustness can be evaluated. In this work we propose an open-source evaluation pipeline, which benchmarks the accuracy of neural embeddings in situations where knowledge graphs may experience semantic and structural changes. We define relation-centric connectivity measures that allow us to connect the link prediction capacity to the structure of the knowledge graph. Such an evaluation pipeline is especially important to simulate the accuracy of embeddings for knowledge graphs that are expected to be frequently updated.
Dividing the Ontology Alignment Task with Semantic Embeddings and Logic-based Modules
Feb 25, 2020



Abstract:Large ontologies still pose serious challenges to state-of-the-art ontology alignment systems. In this paper we present an approach that combines a neural embedding model and logic-based modules to accurately divide an input ontology matching task into smaller and more tractable matching (sub)tasks. We have conducted a comprehensive evaluation using the datasets of the Ontology Alignment Evaluation Initiative. The results are encouraging and suggest that the proposed method is adequate in practice and can be integrated within the workflow of systems unable to cope with very large ontologies.
OpenBioLink: A resource and benchmarking framework for large-scale biomedical link prediction
Dec 10, 2019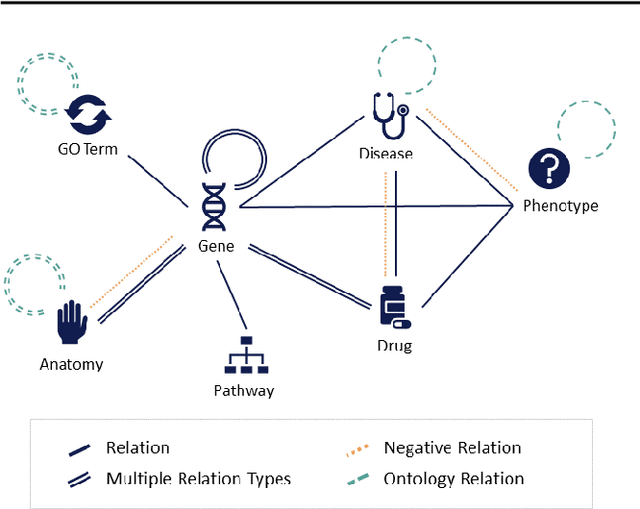
Abstract:SUMMARY: Recently, novel machine-learning algorithms have shown potential for predicting undiscovered links in biomedical knowledge networks. However, dedicated benchmarks for measuring algorithmic progress have not yet emerged. With OpenBioLink, we introduce a large-scale, high-quality and highly challenging biomedical link prediction benchmark to transparently and reproducibly evaluate such algorithms. Furthermore, we present preliminary baseline evaluation results. AVAILABILITY AND IMPLEMENTATION: Source code, data and supplementary files are openly available at https://github.com/OpenBioLink/OpenBioLink CONTACT: matthias.samwald ((at)) meduniwien.ac.at
Global and local evaluation of link prediction tasks with neural embeddings
Jul 27, 2018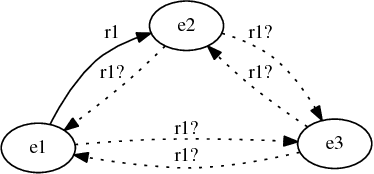
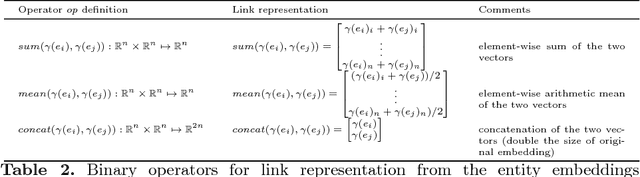
Abstract:We focus our attention on the link prediction problem for knowledge graphs, which is treated herein as a binary classification task on neural embeddings of the entities. By comparing, combining and extending different methodologies for link prediction on graph-based data coming from different domains, we formalize a unified methodology for the quality evaluation benchmark of neural embeddings for knowledge graphs. This benchmark is then used to empirically investigate the potential of training neural embeddings globally for the entire graph, as opposed to the usual way of training embeddings locally for a specific relation. This new way of testing the quality of the embeddings evaluates the performance of binary classifiers for scalable link prediction with limited data. Our evaluation pipeline is made open source, and with this we aim to draw more attention of the community towards an important issue of transparency and reproducibility of the neural embeddings evaluations.
Breaking-down the Ontology Alignment Task with a Lexical Index and Neural Embeddings
May 31, 2018



Abstract:Large ontologies still pose serious challenges to state-of-the-art ontology alignment systems. In the paper we present an approach that combines a lexical index, a neural embedding model and locality modules to effectively divide an input ontology matching task into smaller and more tractable matching (sub)tasks. We have conducted a comprehensive evaluation using the datasets of the Ontology Alignment Evaluation Initiative. The results are encouraging and suggest that the proposed methods are adequate in practice and can be integrated within the workflow of state-of-the-art systems.
Fast and scalable learning of neuro-symbolic representations of biomedical knowledge
Apr 30, 2018



Abstract:In this work we address the problem of fast and scalable learning of neuro-symbolic representations for general biological knowledge. Based on a recently published comprehensive biological knowledge graph (Alshahrani, 2017) that was used for demonstrating neuro-symbolic representation learning, we show how to train fast (under 1 minute) log-linear neural embeddings of the entities. We utilize these representations as inputs for machine learning classifiers to enable important tasks such as biological link prediction. Classifiers are trained by concatenating learned entity embeddings to represent entity relations, and training classifiers on the concatenated embeddings to discern true relations from automatically generated negative examples. Our simple embedding methodology greatly improves on classification error compared to previously published state-of-the-art results, yielding a maximum increase of $+0.28$ F-measure and $+0.22$ ROC AUC scores for the most difficult biological link prediction problem. Finally, our embedding approach is orders of magnitude faster to train ($\leq$ 1 minute vs. hours), much more economical in terms of embedding dimensions ($d=50$ vs. $d=512$), and naturally encodes the directionality of the asymmetric biological relations, that can be controlled by the order with which we concatenate the embeddings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge