Artur Speiser
Teaching deep neural networks to localize sources in super-resolution microscopy by combining simulation-based learning and unsupervised learning
Jun 27, 2019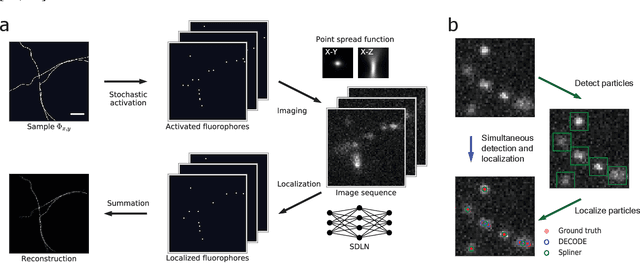
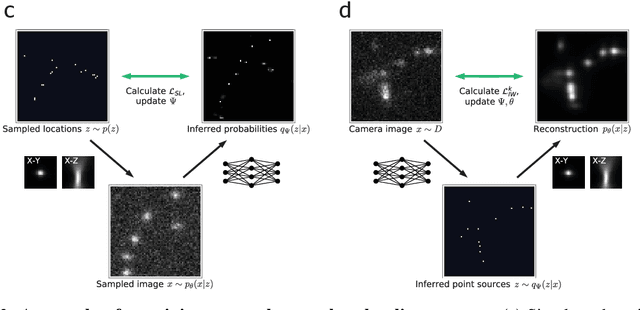

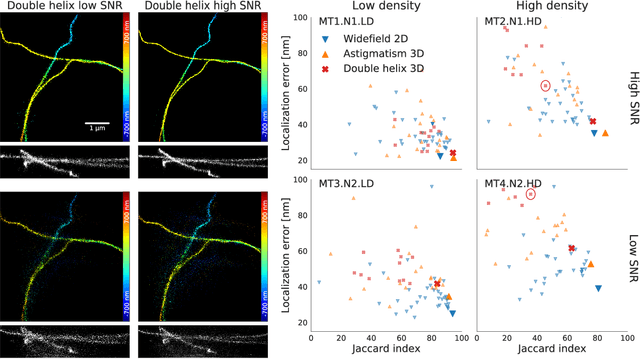
Abstract:Single-molecule localization microscopy constructs super-resolution images by the sequential imaging and computational localization of sparsely activated fluorophores. Accurate and efficient fluorophore localization algorithms are key to the success of this computational microscopy method. We present a novel localization algorithm based on deep learning which significantly improves upon the state of the art. Our contributions are a novel network architecture for simultaneous detection and localization, and a new training algorithm which enables this deep network to solve the Bayesian inverse problem of detecting and localizing single molecules. Our network architecture uses temporal context from multiple sequentially imaged frames to detect and localize molecules. Our training algorithm combines simulation-based supervised learning with autoencoder-based unsupervised learning to make it more robust against mismatch in the generative model. We demonstrate the performance of our method on datasets imaged using a variety of point spread functions and fluorophore densities. While existing localization algorithms can achieve optimal localization accuracy in data with low fluorophore density, they are confounded by high densities. Our method significantly outperforms the state of the art at high densities and thus, enables faster imaging than previous approaches. Our work also more generally shows how to train deep networks to solve challenging Bayesian inverse problems in biology and physics.
Fast amortized inference of neural activity from calcium imaging data with variational autoencoders
Nov 06, 2017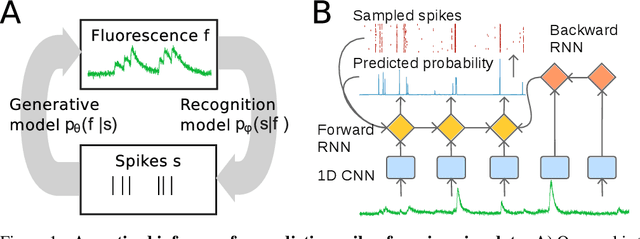
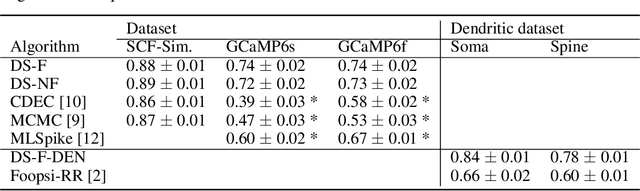
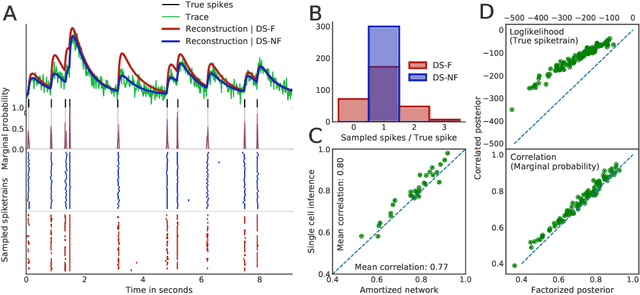
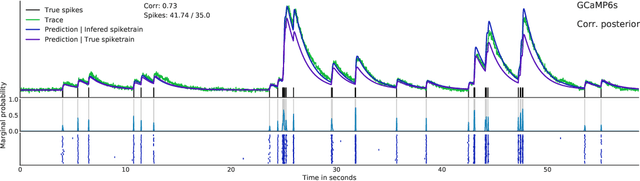
Abstract:Calcium imaging permits optical measurement of neural activity. Since intracellular calcium concentration is an indirect measurement of neural activity, computational tools are necessary to infer the true underlying spiking activity from fluorescence measurements. Bayesian model inversion can be used to solve this problem, but typically requires either computationally expensive MCMC sampling, or faster but approximate maximum-a-posteriori optimization. Here, we introduce a flexible algorithmic framework for fast, efficient and accurate extraction of neural spikes from imaging data. Using the framework of variational autoencoders, we propose to amortize inference by training a deep neural network to perform model inversion efficiently. The recognition network is trained to produce samples from the posterior distribution over spike trains. Once trained, performing inference amounts to a fast single forward pass through the network, without the need for iterative optimization or sampling. We show that amortization can be applied flexibly to a wide range of nonlinear generative models and significantly improves upon the state of the art in computation time, while achieving competitive accuracy. Our framework is also able to represent posterior distributions over spike-trains. We demonstrate the generality of our method by proposing the first probabilistic approach for separating backpropagating action potentials from putative synaptic inputs in calcium imaging of dendritic spines.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge