Anton François
Train-Free Segmentation in MRI with Cubical Persistent Homology
Jan 02, 2024Abstract:We describe a new general method for segmentation in MRI scans using Topological Data Analysis (TDA), offering several advantages over traditional machine learning approaches. It works in three steps, first identifying the whole object to segment via automatic thresholding, then detecting a distinctive subset whose topology is known in advance, and finally deducing the various components of the segmentation. Although convoking classical ideas of TDA, such an algorithm has never been proposed separately from deep learning methods. To achieve this, our approach takes into account, in addition to the homology of the image, the localization of representative cycles, a piece of information that seems never to have been exploited in this context. In particular, it offers the ability to perform segmentation without the need for large annotated data sets. TDA also provides a more interpretable and stable framework for segmentation by explicitly mapping topological features to segmentation components. By adapting the geometric object to be detected, the algorithm can be adjusted to a wide range of data segmentation challenges. We carefully study the examples of glioblastoma segmentation in brain MRI, where a sphere is to be detected, as well as myocardium in cardiac MRI, involving a cylinder, and cortical plate detection in fetal brain MRI, whose 2D slices are circles. We compare our method to state-of-the-art algorithms.
A deep residual learning implementation of Metamorphosis
Feb 01, 2022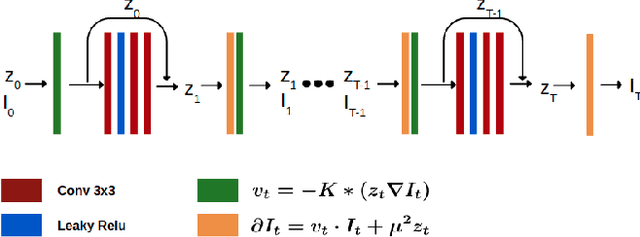
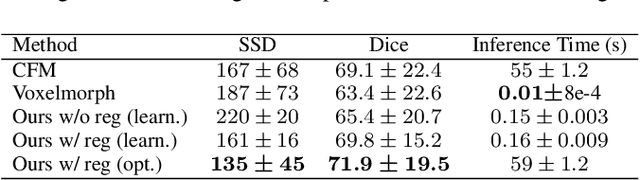
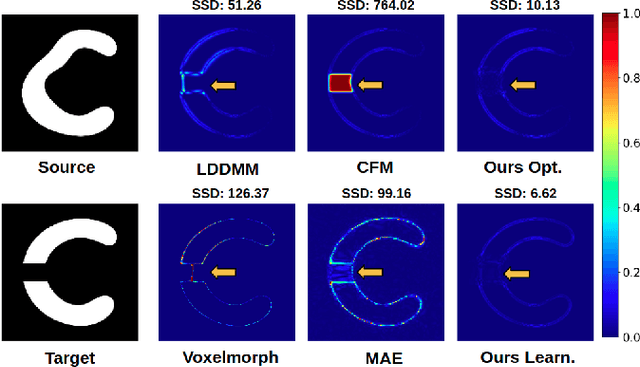
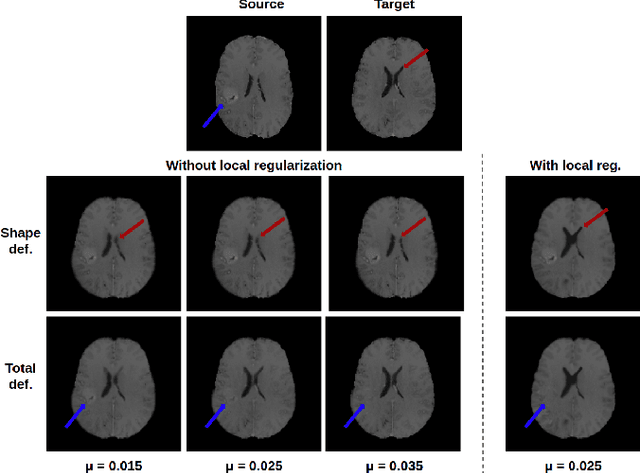
Abstract:In medical imaging, most of the image registration methods implicitly assume a one-to-one correspondence between the source and target images (i.e., diffeomorphism). However, this is not necessarily the case when dealing with pathological medical images (e.g., presence of a tumor, lesion, etc.). To cope with this issue, the Metamorphosis model has been proposed. It modifies both the shape and the appearance of an image to deal with the geometrical and topological differences. However, the high computational time and load have hampered its applications so far. Here, we propose a deep residual learning implementation of Metamorphosis that drastically reduces the computational time at inference. Furthermore, we also show that the proposed framework can easily integrate prior knowledge of the localization of topological changes (e.g., segmentation masks) that can act as spatial regularization to correctly disentangle appearance and shape changes. We test our method on the BraTS 2021 dataset, showing that it outperforms current state-of-the-art methods in the alignment of images with brain tumors.
Metamorphic image registration using a semi-Lagrangian scheme
Jun 16, 2021

Abstract:In this paper, we propose an implementation of both Large Deformation Diffeomorphic Metric Mapping (LDDMM) and Metamorphosis image registration using a semi-Lagrangian scheme for geodesic shooting. We propose to solve both problems as an inexact matching providing a single and unifying cost function. We demonstrate that for image registration the use of a semi-Lagrangian scheme is more stable than a standard Eulerian scheme. Our GPU implementation is based on PyTorch, which greatly simplifies and accelerates the computations thanks to its powerful automatic differentiation engine. It will be freely available at https://github.com/antonfrancois/Demeter_metamorphosis.
* SEE GSI 2021
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge