Annalisa Barla
The effect of data augmentation and 3D-CNN depth on Alzheimer's Disease detection
Sep 13, 2023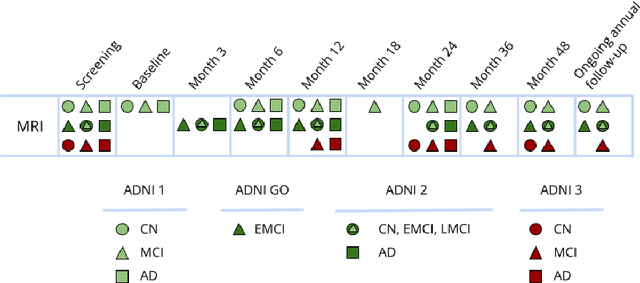
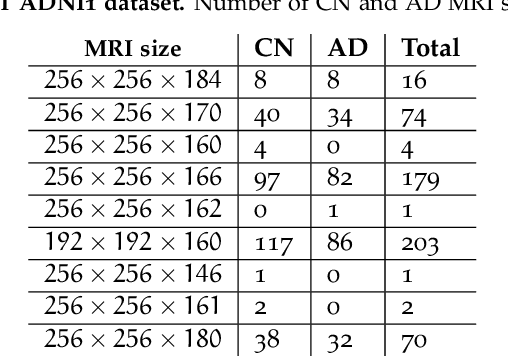
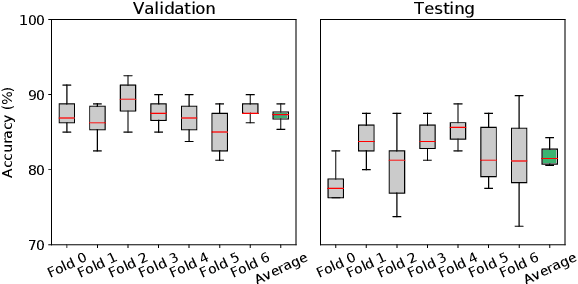
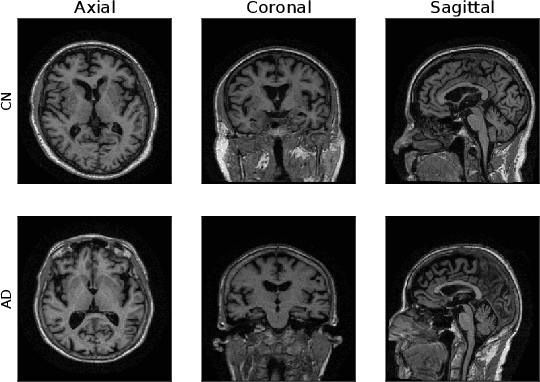
Abstract:Machine Learning (ML) has emerged as a promising approach in healthcare, outperforming traditional statistical techniques. However, to establish ML as a reliable tool in clinical practice, adherence to best practices regarding data handling, experimental design, and model evaluation is crucial. This work summarizes and strictly observes such practices to ensure reproducible and reliable ML. Specifically, we focus on Alzheimer's Disease (AD) detection, which serves as a paradigmatic example of challenging problem in healthcare. We investigate the impact of different data augmentation techniques and model complexity on the overall performance. We consider MRI data from ADNI dataset to address a classification problem employing 3D Convolutional Neural Network (CNN). The experiments are designed to compensate for data scarcity and initial random parameters by utilizing cross-validation and multiple training trials. Within this framework, we train 15 predictive models, considering three different data augmentation strategies and five distinct 3D CNN architectures, each varying in the number of convolutional layers. Specifically, the augmentation strategies are based on affine transformations, such as zoom, shift, and rotation, applied concurrently or separately. The combined effect of data augmentation and model complexity leads to a variation in prediction performance up to 10% of accuracy. When affine transformation are applied separately, the model is more accurate, independently from the adopted architecture. For all strategies, the model accuracy followed a concave behavior at increasing number of convolutional layers, peaking at an intermediate value of layers. The best model (8 CL, (B)) is the most stable across cross-validation folds and training trials, reaching excellent performance both on the testing set and on an external test set.
Group induced graphical lasso allows for discovery of molecular pathways-pathways interactions
Nov 21, 2018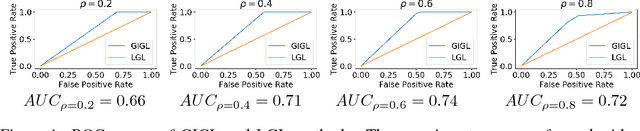
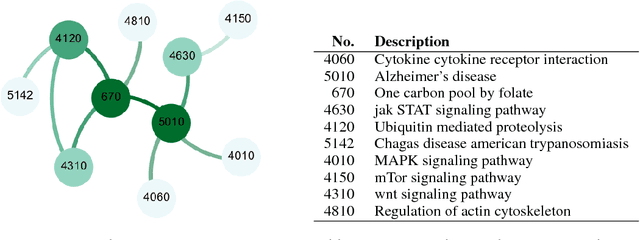
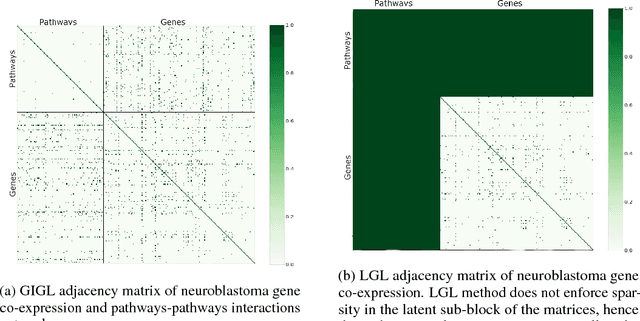
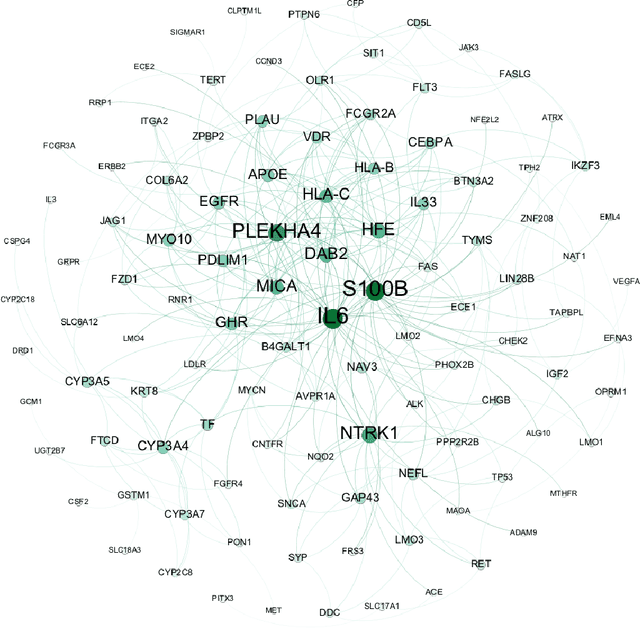
Abstract:Complex systems may contain heterogeneous types of variables that interact in a multi-level and multi-scale manner. In this context, high-level layers may considered as groups of variables interacting in lower-level layers. This is particularly true in biology, where, for example, genes are grouped in pathways and two types of interactions are present: pathway-pathway interactions and gene-gene interactions. However, from data it is only possible to measure the expression of genes while it is impossible to directly measure the activity of pathways. Nevertheless, the knowledge on the inter-dependence between the groups and the variables allows for a multi-layer network inference, on both observed variables and groups, even if no direct information on the latter is present in the data (hence groups are considered as latent). In this paper, we propose an extension of the latent graphical lasso method that leverages on the knowledge of the inter-links between the hidden (groups) and observed layers. The method exploits the knowledge of group structure that influence the behaviour of observed variables to retrieve a two layers network. Its efficacy was tested on synthetic data to check its ability in retrieving the network structure compared to the ground truth. We present a case study on Neuroblastoma, which shows how our multi-level inference is relevant in real contexts to infer biologically meaningful connections.
A temporal model for multiple sclerosis course evolution
Dec 02, 2016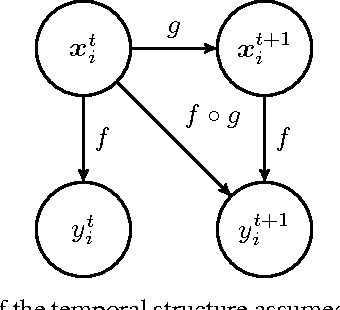

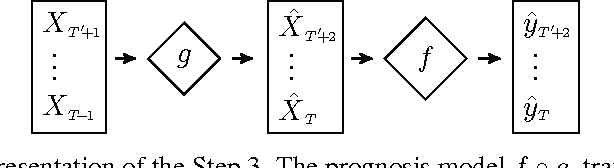
Abstract:Multiple Sclerosis is a degenerative condition of the central nervous system that affects nearly 2.5 million of individuals in terms of their physical, cognitive, psychological and social capabilities. Researchers are currently investigating on the use of patient reported outcome measures for the assessment of impact and evolution of the disease on the life of the patients. To date, a clear understanding on the use of such measures to predict the evolution of the disease is still lacking. In this work we resort to regularized machine learning methods for binary classification and multiple output regression. We propose a pipeline that can be used to predict the disease progression from patient reported measures. The obtained model is tested on a data set collected from an ongoing clinical research project.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge