Anika Grüneboom
EfficientBioAI: Making Bioimaging AI Models Efficient in Energy, Latency and Representation
Jun 09, 2023Abstract:Artificial intelligence (AI) has been widely used in bioimage image analysis nowadays, but the efficiency of AI models, like the energy consumption and latency is not ignorable due to the growing model size and complexity, as well as the fast-growing analysis needs in modern biomedical studies. Like we can compress large images for efficient storage and sharing, we can also compress the AI models for efficient applications and deployment. In this work, we present EfficientBioAI, a plug-and-play toolbox that can compress given bioimaging AI models for them to run with significantly reduced energy cost and inference time on both CPU and GPU, without compromise on accuracy. In some cases, the prediction accuracy could even increase after compression, since the compression procedure could remove redundant information in the model representation and therefore reduce over-fitting. From four different bioimage analysis applications, we observed around 2-5 times speed-up during inference and 30-80$\%$ saving in energy. Cutting the runtime of large scale bioimage analysis from days to hours or getting a two-minutes bioimaging AI model inference done in near real-time will open new doors for method development and biomedical discoveries. We hope our toolbox will facilitate resource-constrained bioimaging AI and accelerate large-scale AI-based quantitative biological studies in an eco-friendly way, as well as stimulate further research on the efficiency of bioimaging AI.
SYNTA: A novel approach for deep learning-based image analysis in muscle histopathology using photo-realistic synthetic data
Aug 03, 2022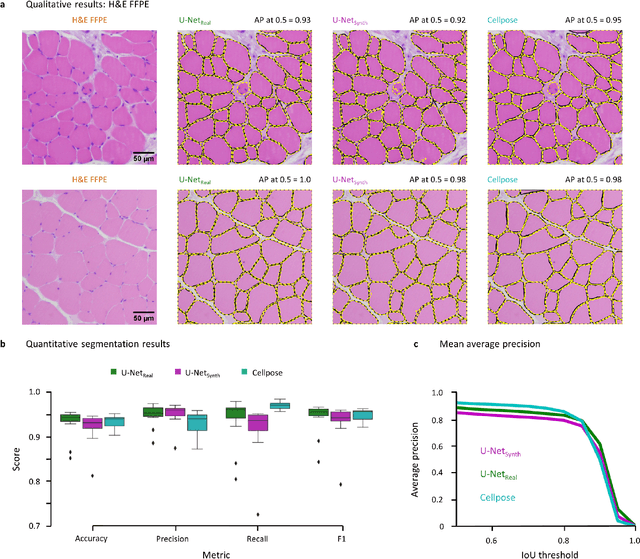
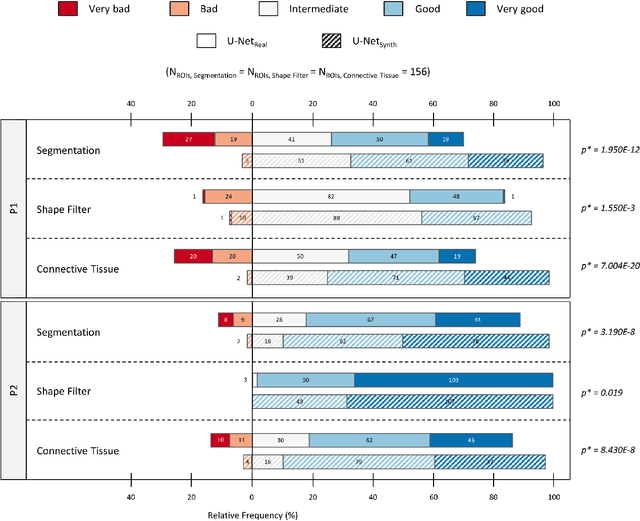
Abstract:Artificial intelligence (AI), machine learning, and deep learning (DL) methods are becoming increasingly important in the field of biomedical image analysis. However, to exploit the full potential of such methods, a representative number of experimentally acquired images containing a significant number of manually annotated objects is needed as training data. Here we introduce SYNTA (synthetic data) as a novel approach for the generation of synthetic, photo-realistic, and highly complex biomedical images as training data for DL systems. We show the versatility of our approach in the context of muscle fiber and connective tissue analysis in histological sections. We demonstrate that it is possible to perform robust and expert-level segmentation tasks on previously unseen real-world data, without the need for manual annotations using synthetic training data alone. Being a fully parametric technique, our approach poses an interpretable and controllable alternative to Generative Adversarial Networks (GANs) and has the potential to significantly accelerate quantitative image analysis in a variety of biomedical applications in microscopy and beyond.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge