Andy Schuetz
Medical Concept Representation Learning from Electronic Health Records and its Application on Heart Failure Prediction
Jun 20, 2017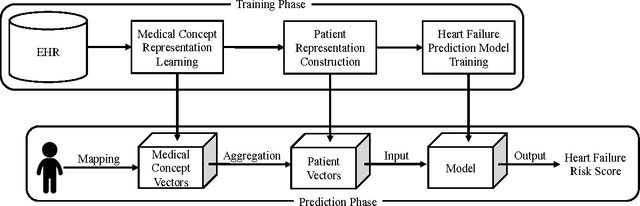
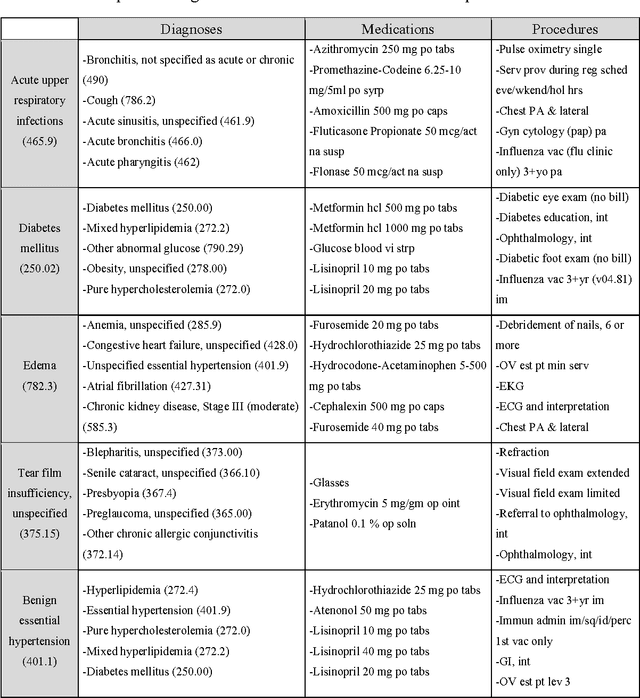
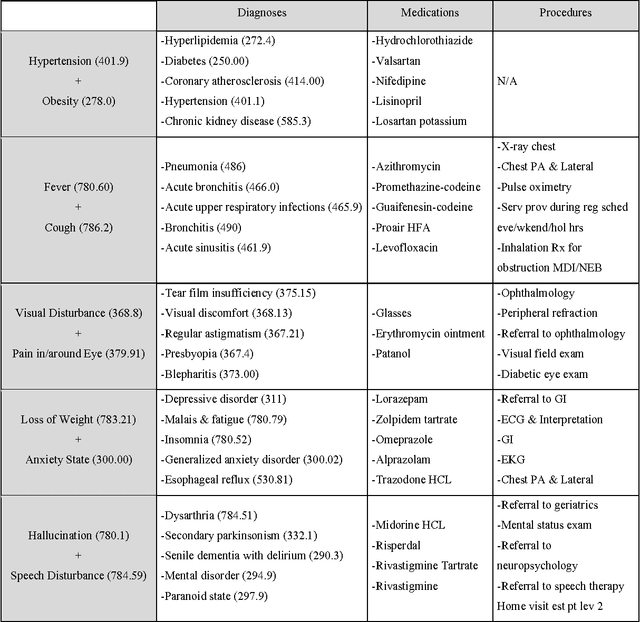
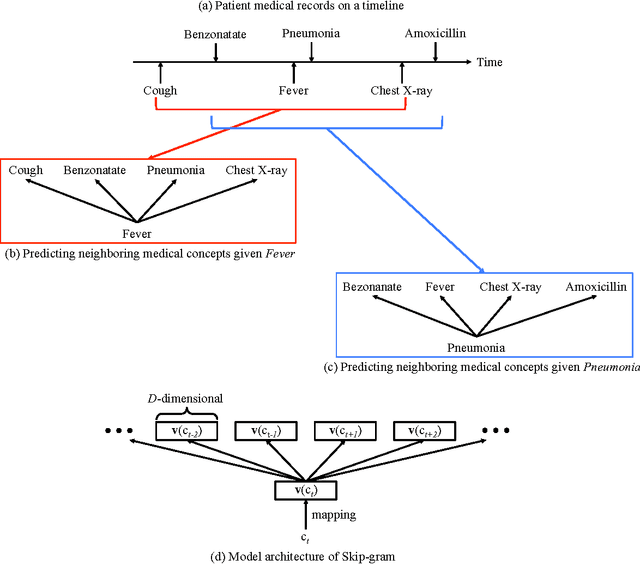
Abstract:Objective: To transform heterogeneous clinical data from electronic health records into clinically meaningful constructed features using data driven method that rely, in part, on temporal relations among data. Materials and Methods: The clinically meaningful representations of medical concepts and patients are the key for health analytic applications. Most of existing approaches directly construct features mapped to raw data (e.g., ICD or CPT codes), or utilize some ontology mapping such as SNOMED codes. However, none of the existing approaches leverage EHR data directly for learning such concept representation. We propose a new way to represent heterogeneous medical concepts (e.g., diagnoses, medications and procedures) based on co-occurrence patterns in longitudinal electronic health records. The intuition behind the method is to map medical concepts that are co-occuring closely in time to similar concept vectors so that their distance will be small. We also derive a simple method to construct patient vectors from the related medical concept vectors. Results: For qualitative evaluation, we study similar medical concepts across diagnosis, medication and procedure. In quantitative evaluation, our proposed representation significantly improves the predictive modeling performance for onset of heart failure (HF), where classification methods (e.g. logistic regression, neural network, support vector machine and K-nearest neighbors) achieve up to 23% improvement in area under the ROC curve (AUC) using this proposed representation. Conclusion: We proposed an effective method for patient and medical concept representation learning. The resulting representation can map relevant concepts together and also improves predictive modeling performance.
RETAIN: An Interpretable Predictive Model for Healthcare using Reverse Time Attention Mechanism
Feb 26, 2017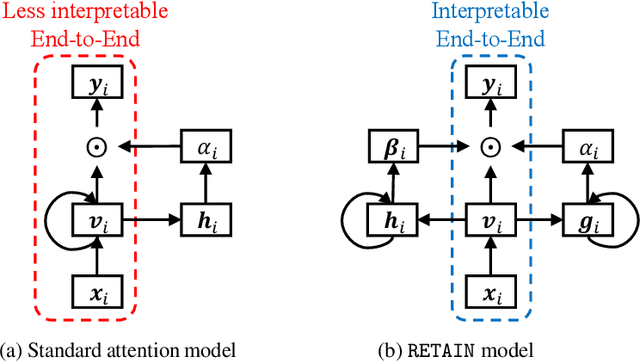

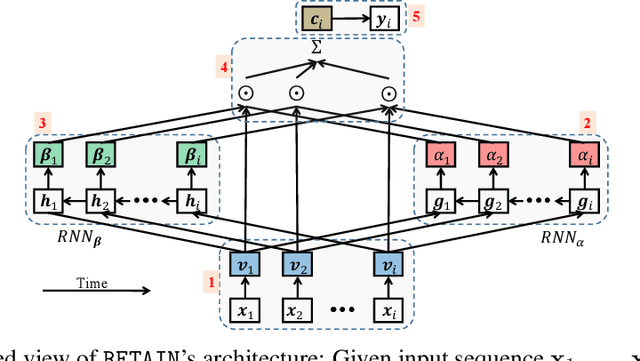
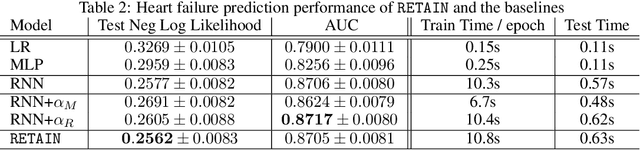
Abstract:Accuracy and interpretability are two dominant features of successful predictive models. Typically, a choice must be made in favor of complex black box models such as recurrent neural networks (RNN) for accuracy versus less accurate but more interpretable traditional models such as logistic regression. This tradeoff poses challenges in medicine where both accuracy and interpretability are important. We addressed this challenge by developing the REverse Time AttentIoN model (RETAIN) for application to Electronic Health Records (EHR) data. RETAIN achieves high accuracy while remaining clinically interpretable and is based on a two-level neural attention model that detects influential past visits and significant clinical variables within those visits (e.g. key diagnoses). RETAIN mimics physician practice by attending the EHR data in a reverse time order so that recent clinical visits are likely to receive higher attention. RETAIN was tested on a large health system EHR dataset with 14 million visits completed by 263K patients over an 8 year period and demonstrated predictive accuracy and computational scalability comparable to state-of-the-art methods such as RNN, and ease of interpretability comparable to traditional models.
Doctor AI: Predicting Clinical Events via Recurrent Neural Networks
Sep 28, 2016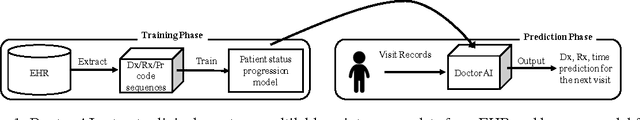
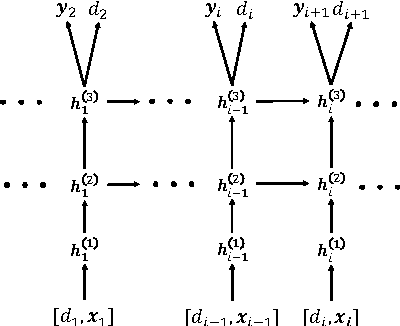
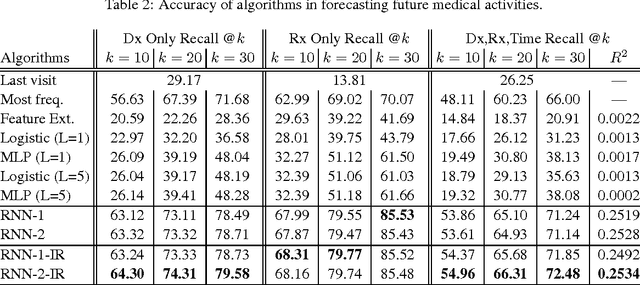
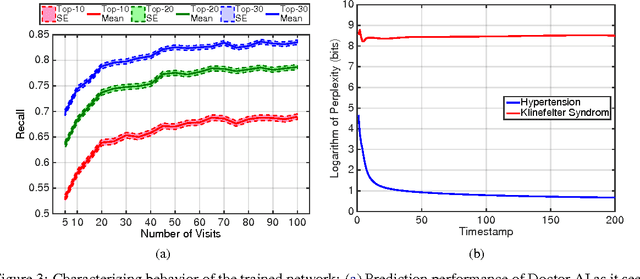
Abstract:Leveraging large historical data in electronic health record (EHR), we developed Doctor AI, a generic predictive model that covers observed medical conditions and medication uses. Doctor AI is a temporal model using recurrent neural networks (RNN) and was developed and applied to longitudinal time stamped EHR data from 260K patients over 8 years. Encounter records (e.g. diagnosis codes, medication codes or procedure codes) were input to RNN to predict (all) the diagnosis and medication categories for a subsequent visit. Doctor AI assesses the history of patients to make multilabel predictions (one label for each diagnosis or medication category). Based on separate blind test set evaluation, Doctor AI can perform differential diagnosis with up to 79% recall@30, significantly higher than several baselines. Moreover, we demonstrate great generalizability of Doctor AI by adapting the resulting models from one institution to another without losing substantial accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge