Andrew Xie
Neural Inverse Rendering from Propagating Light
Jun 05, 2025Abstract:We present the first system for physically based, neural inverse rendering from multi-viewpoint videos of propagating light. Our approach relies on a time-resolved extension of neural radiance caching -- a technique that accelerates inverse rendering by storing infinite-bounce radiance arriving at any point from any direction. The resulting model accurately accounts for direct and indirect light transport effects and, when applied to captured measurements from a flash lidar system, enables state-of-the-art 3D reconstruction in the presence of strong indirect light. Further, we demonstrate view synthesis of propagating light, automatic decomposition of captured measurements into direct and indirect components, as well as novel capabilities such as multi-view time-resolved relighting of captured scenes.
Deep Learning-Based Correction and Unmixing of Hyperspectral Images for Brain Tumor Surgery
Feb 06, 2024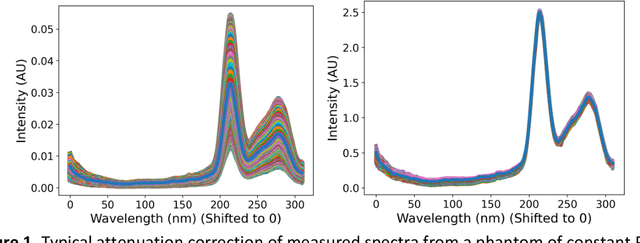
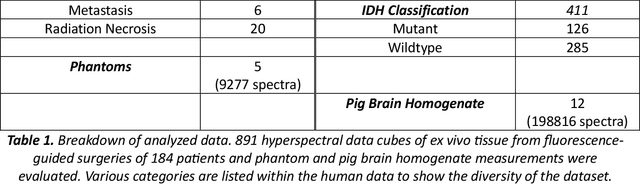
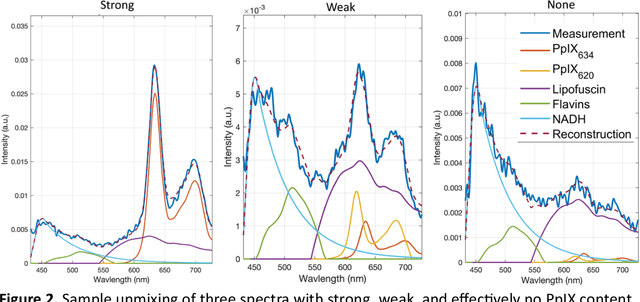
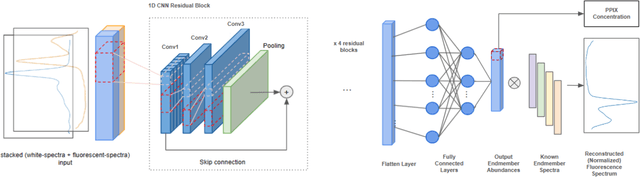
Abstract:Hyperspectral Imaging (HSI) for fluorescence-guided brain tumor resection enables visualization of differences between tissues that are not distinguishable to humans. This augmentation can maximize brain tumor resection, improving patient outcomes. However, much of the processing in HSI uses simplified linear methods that are unable to capture the non-linear, wavelength-dependent phenomena that must be modeled for accurate recovery of fluorophore abundances. We therefore propose two deep learning models for correction and unmixing, which can account for the nonlinear effects and produce more accurate estimates of abundances. Both models use an autoencoder-like architecture to process the captured spectra. One is trained with protoporphyrin IX (PpIX) concentration labels. The other undergoes semi-supervised training, first learning hyperspectral unmixing self-supervised and then learning to correct fluorescence emission spectra for heterogeneous optical and geometric properties using a reference white-light reflectance spectrum in a few-shot manner. The models were evaluated against phantom and pig brain data with known PpIX concentration; the supervised model achieved Pearson correlation coefficients (R values) between the known and computed PpIX concentrations of 0.997 and 0.990, respectively, whereas the classical approach achieved only 0.93 and 0.82. The semi-supervised approach's R values were 0.98 and 0.91, respectively. On human data, the semi-supervised model gives qualitatively more realistic results than the classical method, better removing bright spots of specular reflectance and reducing the variance in PpIX abundance over biopsies that should be relatively homogeneous. These results show promise for using deep learning to improve HSI in fluorescence-guided neurosurgery.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge