Anders Persson
Creating and Benchmarking a Synthetic Dataset for Cloud Optical Thickness Estimation
Nov 23, 2023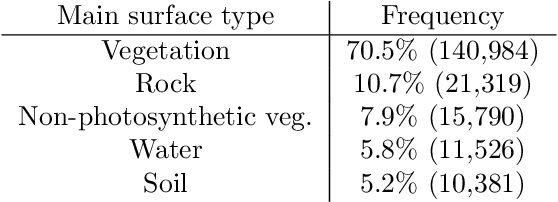
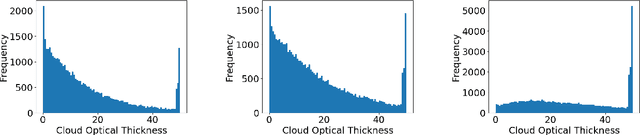
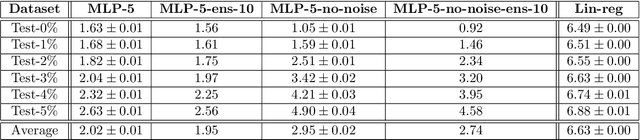
Abstract:Cloud formations often obscure optical satellite-based monitoring of the Earth's surface, thus limiting Earth observation (EO) activities such as land cover mapping, ocean color analysis, and cropland monitoring. The integration of machine learning (ML) methods within the remote sensing domain has significantly improved performance on a wide range of EO tasks, including cloud detection and filtering, but there is still much room for improvement. A key bottleneck is that ML methods typically depend on large amounts of annotated data for training, which is often difficult to come by in EO contexts. This is especially true for the task of cloud optical thickness (COT) estimation. A reliable estimation of COT enables more fine-grained and application-dependent control compared to using pre-specified cloud categories, as is commonly done in practice. To alleviate the COT data scarcity problem, in this work we propose a novel synthetic dataset for COT estimation, where top-of-atmosphere radiances have been simulated for 12 of the spectral bands of the Multi-Spectral Instrument (MSI) sensor onboard Sentinel-2 platforms. These data points have been simulated under consideration of different cloud types, COTs, and ground surface and atmospheric profiles. Extensive experimentation of training several ML models to predict COT from the measured reflectivity of the spectral bands demonstrates the usefulness of our proposed dataset. Generalization to real data is also demonstrated on two satellite image datasets -- one that is publicly available, and one which we have collected and annotated. The synthetic data, the newly collected real dataset, code and models have been made publicly available at https://github.com/aleksispi/ml-cloud-opt-thick.
Faster 3D cardiac CT segmentation with Vision Transformers
Oct 13, 2023Abstract:Accurate segmentation of the heart is essential for personalized blood flow simulations and surgical intervention planning. A recent advancement in image recognition is the Vision Transformer (ViT), which expands the field of view to encompass a greater portion of the global image context. We adapted ViT for three-dimensional volume inputs. Cardiac computed tomography (CT) volumes from 39 patients, featuring up to 20 timepoints representing the complete cardiac cycle, were utilized. Our network incorporates a modified ResNet50 block as well as a ViT block and employs cascade upsampling with skip connections. Despite its increased model complexity, our hybrid Transformer-Residual U-Net framework, termed TRUNet, converges in significantly less time than residual U-Net while providing comparable or superior segmentations of the left ventricle, left atrium, left atrial appendage, ascending aorta, and pulmonary veins. TRUNet offers more precise vessel boundary segmentation and better captures the heart's overall anatomical structure compared to residual U-Net, as confirmed by the absence of extraneous clusters of missegmented voxels. In terms of both performance and training speed, TRUNet exceeded U-Net, a commonly used segmentation architecture, making it a promising tool for 3D semantic segmentation tasks in medical imaging. The code for TRUNet is available at github.com/ljollans/TRUNet.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge