Anaïs Baudot
MODIS: Multi-Omics Data Integration for Small and Unpaired Datasets
Mar 24, 2025Abstract:A key challenge today lies in the ability to efficiently handle multi-omics data since such multimodal data may provide a more comprehensive overview of the underlying processes in a system. Yet it comes with challenges: multi-omics data are most often unpaired and only partially labeled, moreover only small amounts of data are available in some situation such as rare diseases. We propose MODIS which stands for Multi-Omics Data Integration for Small and unpaired datasets, a semi supervised approach to account for these particular settings. MODIS learns a probabilistic coupling of heterogeneous data modalities and learns a shared latent space where modalities are aligned. We rely on artificial data to build controlled experiments to explore how much supervision is needed for an accurate alignment of modalities, and how our approach enables dealing with new conditions for which few data are available. The code is available athttps://github.com/VILLOUTREIXLab/MODIS.
Zoo Guide to Network Embedding
May 05, 2023Abstract:Networks have provided extremely successful models of data and complex systems. Yet, as combinatorial objects, networks do not have in general intrinsic coordinates and do not typically lie in an ambient space. The process of assigning an embedding space to a network has attracted lots of interest in the past few decades, and has been efficiently applied to fundamental problems in network inference, such as link prediction, node classification, and community detection. In this review, we provide a user-friendly guide to the network embedding literature and current trends in this field which will allow the reader to navigate through the complex landscape of methods and approaches emerging from the vibrant research activity on these subjects.
Universal Multilayer Network Exploration by Random Walk with Restart
Jul 09, 2021
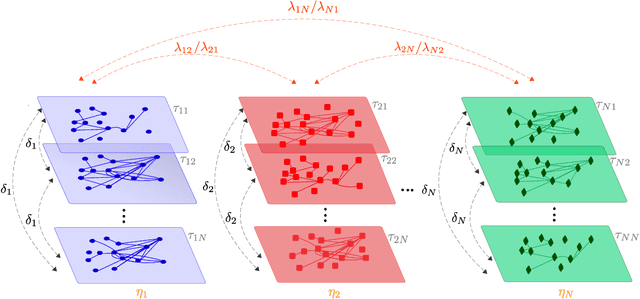

Abstract:The amount and variety of data is increasing drastically for several years. These data are often represented as networks, which are then explored with approaches arising from network theory. Recent years have witnessed the extension of network exploration methods to leverage more complex and richer network frameworks. Random walks, for instance, have been extended to explore multilayer networks. However, current random walk approaches are limited in the combination and heterogeneity of network layers they can handle. New analytical and numerical random walk methods are needed to cope with the increasing diversity and complexity of multilayer networks. We propose here MultiXrank, a Python package that enables Random Walk with Restart (RWR) on any kind of multilayer network with an optimized implementation. This package is supported by a universal mathematical formulation of the RWR. We evaluated MultiXrank with leave-one-out cross-validation and link prediction, and introduced protocols to measure the impact of the addition or removal of multilayer network data on prediction performances. We further measured the sensitivity of MultiXrank to input parameters by in-depth exploration of the parameter space. Finally, we illustrate the versatility of MultiXrank with different use-cases of unsupervised node prioritization and supervised classification in the context of human genetic diseases.
MultiVERSE: a multiplex and multiplex-heterogeneous network embedding approach
Aug 23, 2020



Abstract:Network embedding approaches are gaining momentum to analyse a large variety of networks. Indeed, these approaches have demonstrated their efficiency for tasks such as community detection, node classification, and link prediction. However, very few network embedding methods have been specifically designed to handle multiplex networks, i.e. networks composed of different layers sharing the same set of nodes but having different types of edges. Moreover, to our knowledge, existing approaches cannot embed multiple nodes from multiplex-heterogeneous networks, i.e. networks composed of several layers containing both different types of nodes and edges. In this study, we propose MultiVERSE, an extension of the VERSE method with Random Walks with Restart on Multiplex (RWR-M) and Multiplex-Heterogeneous (RWR-MH) networks. MultiVERSE is a fast and scalable method to learn node embeddings from multiplex and multiplex-heterogeneous networks. We evaluate MultiVERSE on several biological and social networks and demonstrate its efficiency. MultiVERSE indeed outperforms most of the other methods in the tasks of link prediction and network reconstruction for multiplex network embedding, and is also efficient in the task of link prediction for multiplex-heterogeneous network embedding. Finally, we apply MultiVERSE to study rare disease-gene associations using link prediction and clustering. MultiVERSE is freely available on github at https://github.com/Lpiol/MultiVERSE.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge