Alycia Lee
Beyond Scale: the Diversity Coefficient as a Data Quality Metric Demonstrates LLMs are Pre-trained on Formally Diverse Data
Jun 24, 2023Abstract:Current trends to pre-train capable Large Language Models (LLMs) mostly focus on scaling of model and dataset size. However, the quality of pre-training data is an important factor for training powerful LLMs, yet it is a nebulous concept that has not been fully characterized. Therefore, we use the recently proposed Task2Vec diversity coefficient to ground and understand formal aspects of data quality, to go beyond scale alone. Specifically, we measure the diversity coefficient of publicly available pre-training datasets to demonstrate that their formal diversity is high when compared to theoretical lower and upper bounds. In addition, to build confidence in the diversity coefficient, we conduct interpretability experiments and find that the coefficient aligns with intuitive properties of diversity, e.g., it increases as the number of latent concepts increases. We conclude the diversity coefficient is reliable, show it's high for publicly available LLM datasets, and conjecture it can be used to build useful diverse datasets for LLMs.
Batched Stochastic Bayesian Optimization via Combinatorial Constraints Design
Apr 17, 2019
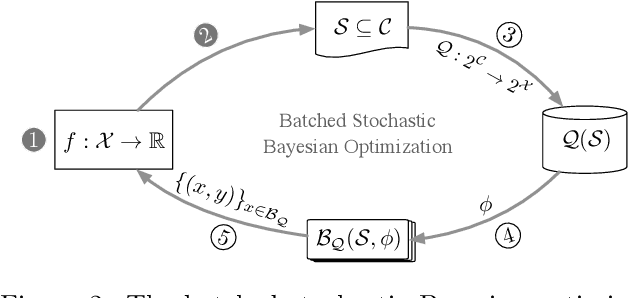
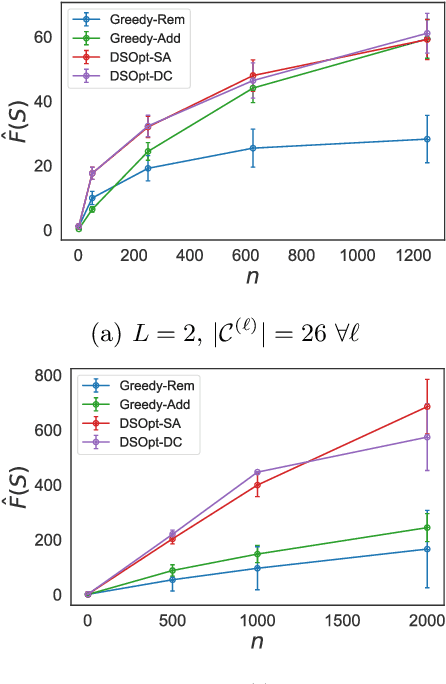
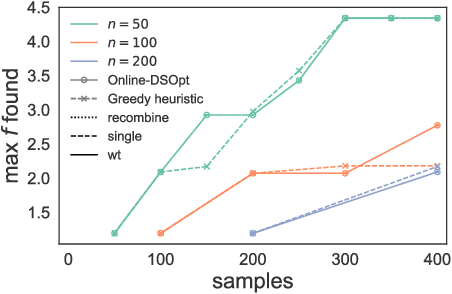
Abstract:In many high-throughput experimental design settings, such as those common in biochemical engineering, batched queries are more cost effective than one-by-one sequential queries. Furthermore, it is often not possible to directly choose items to query. Instead, the experimenter specifies a set of constraints that generates a library of possible items, which are then selected stochastically. Motivated by these considerations, we investigate \emph{Batched Stochastic Bayesian Optimization} (BSBO), a novel Bayesian optimization scheme for choosing the constraints in order to guide exploration towards items with greater utility. We focus on \emph{site-saturation mutagenesis}, a prototypical setting of BSBO in biochemical engineering, and propose a natural objective function for this problem. Importantly, we show that our objective function can be efficiently decomposed as a difference of submodular functions (DS), which allows us to employ DS optimization tools to greedily identify sets of constraints that increase the likelihood of finding items with high utility. Our experimental results show that our algorithm outperforms common heuristics on both synthetic and two real protein datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge